| IDH3G | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | IDH3G, H-IDHG, isocitrate dehydrogenase 3 (NAD(+)) gamma, isocitrate dehydrogenase (NAD(+)) 3 gamma, isocitrate dehydrogenase (NAD(+)) 3 non-catalytic subunit gamma | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 300089; MGI: 1099463; HomoloGene: 55803; GeneCards: IDH3G; OMA:IDH3G - orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Isocitrate dehydrogenase subunit gamma, mitochondrial is an enzyme that in humans is encoded by the IDH3G gene.
Isocitrate dehydrogenases (IDHs) catalyze the oxidative decarboxylation of isocitrate to 2-oxoglutarate. These enzymes belong to two distinct subclasses, one of which utilizes NAD(+) as the electron acceptor and the other NADP(+). Five isocitrate dehydrogenases have been reported: three NAD(+)-dependent isocitrate dehydrogenases, which localize to the mitochondrial matrix, and two NADP(+)-dependent isocitrate dehydrogenases, one of which is mitochondrial and the other predominantly cytosolic. NAD(+)-dependent isocitrate dehydrogenases catalyze the allosterically regulated rate-limiting step of the tricarboxylic acid cycle. Each isozyme is a heterotetramer that is composed of two alpha subunits, one beta subunit, and one gamma subunit. The protein encoded by this gene is the gamma subunit of one isozyme of NAD(+)-dependent isocitrate dehydrogenase. This gene is a candidate gene for periventricular heterotopia. Several alternatively spliced transcript variants of this gene have been described, but only some of their full length natures have been determined.
Structure
IDH3 is one of three isocitrate dehydrogenase isozymes, the other two being IDH1 and IDH2, and encoded by one of five isocitrate dehydrogenase genes, which are IDH1, IDH2, IDH3A, IDH3B, and IDH3G. The genes IDH3A, IDH3B, and IDH3G encode subunits of IDH3, which is a heterotetramer composed of two 37-kDa α subunits (IDH3α), one 39-kDa β subunit (IDH3β), and one 39-kDa γ subunit (IDH3γ), each with distinct isoelectric points. Alignment of their amino acid sequences reveals ~40% identity between IDH3α and IDH3β, ~42% identity between IDH3α and IDH3γ, and an even closer identity of 53% between IDH3β and IDH3γ, for an overall 34% identity and 23% similarity across all three subunit types. Notably, Arg88 in IDH3α is essential for IDH3 catalytic activity, whereas the equivalent Arg99 in IDH3β and Arg97 in IDH3γ are largely involved in the enzyme's allosteric regulation by ADP and NAD. Thus, it is possible that these subunits arose from gene duplication of a common ancestral gene, and the original catalytic Arg residue were adapted to allosteric functions in the β- and γ-subunits. Likewise, Asp181 in IDH3α is essential for catalysis, while the equivalent Asp192 in IDH3β and Asp190 in IDH3γ enhance NAD- and Mn-binding. Since the oxidative decarboxylation catalyzed by IDH3 requires binding of NAD, Mn, and the substrate isocitrate, all three subunits participate in the catalytic reaction. Moreover, studies of the enzyme in pig heart reveal that the αβ and αγ dimers constitute two binding sites for each of its ligands, including isocitrate, Mn, and NAD, in one IDH3 tetramer.
Function
As an isocitrate dehydrogenase, IDH3 catalyzes the irreversible oxidative decarboxylation of isocitrate to yield α-ketoglutarate (α-KG) and CO2 as part of the TCA cycle in glucose metabolism. This step also allows for the concomitant reduction of NAD+ to NADH, which is then used to generate ATP through the electron transport chain. Notably, IDH3 relies on NAD+ as its electron acceptor, as opposed to NADP+ like IDH1 and IDH2. IDH3 activity is regulated by the energy needs of the cell: when the cell requires energy, IDH3 is activated by ADP; and when energy is no longer required, IDH3 is inhibited by ATP and NADH. This allosteric regulation allows IDH3 to function as a rate-limiting step in the TCA cycle. Within cells, IDH3 and its subunits have been observed to localize to the mitochondria.
Clinical Significance
The IDH3G gene may be involved in drug resistance in gastric cancer.
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles.
[[File: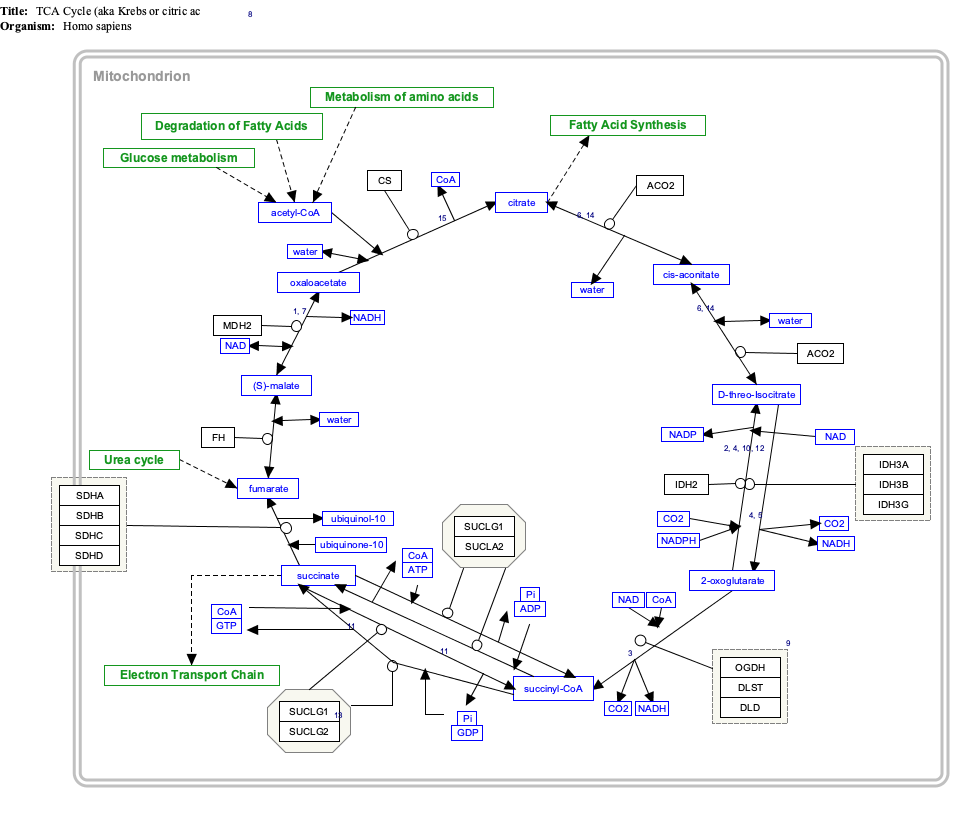

- The interactive pathway map can be edited at WikiPathways: "TCACycle_WP78".
See also
References
- ^ GRCh38: Ensembl release 89: ENSG00000067829 – Ensembl, May 2017
- ^ GRCm38: Ensembl release 89: ENSMUSG00000002010 – Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Brenner V, Nyakatura G, Rosenthal A, Platzer M (August 1997). "Genomic organization of two novel genes on human Xq28: compact head to head arrangement of IDH gamma and TRAP delta is conserved in rat and mouse". Genomics. 44 (1): 8–14. doi:10.1006/geno.1997.4822. PMID 9286695.
- ^ "Entrez Gene: IDH3G isocitrate dehydrogenase 3 (NAD+) gamma".
- Dimitrov L, Hong CS, Yang C, Zhuang Z, Heiss JD (2015). "New developments in the pathogenesis and therapeutic targeting of the IDH1 mutation in glioma". International Journal of Medical Sciences. 12 (3): 201–213. doi:10.7150/ijms.11047. PMC 4323358. PMID 25678837.
- ^ Zeng L, Morinibu A, Kobayashi M, Zhu Y, Wang X, Goto Y, et al. (September 2015). "Aberrant IDH3α expression promotes malignant tumor growth by inducing HIF-1-mediated metabolic reprogramming and angiogenesis". Oncogene. 34 (36): 4758–4766. doi:10.1038/onc.2014.411. PMID 25531325.
- ^ Bzymek KP, Colman RF (May 2007). "Role of alpha-Asp181, beta-Asp192, and gamma-Asp190 in the distinctive subunits of human NAD-specific isocitrate dehydrogenase". Biochemistry. 46 (18): 5391–5397. doi:10.1021/bi700061t. PMID 17432878.
- ^ Soundar S, O'Hagan M, Fomulu KS, Colman RF (July 2006). "Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent isocitrate dehydrogenase". The Journal of Biological Chemistry. 281 (30): 21073–21081. doi:10.1074/jbc.m602956200. PMID 16737955.
- ^ Soundar S, Park JH, Huh TL, Colman RF (December 2003). "Evaluation by mutagenesis of the importance of 3 arginines in alpha, beta, and gamma subunits of human NAD-dependent isocitrate dehydrogenase". The Journal of Biological Chemistry. 278 (52): 52146–52153. doi:10.1074/jbc.m306178200. PMID 14555658.
- Dange M, Colman RF (July 2010). "Each conserved active site tyr in the three subunits of human isocitrate dehydrogenase has a different function". The Journal of Biological Chemistry. 285 (27): 20520–20525. doi:10.1074/jbc.m110.115386. PMC 2898308. PMID 20435888.
- ^ Huh TL, Kim YO, Oh IU, Song BJ, Inazawa J (March 1996). "Assignment of the human mitochondrial NAD+ -specific isocitrate dehydrogenase alpha subunit (IDH3A) gene to 15q25.1-->q25.2by in situ hybridization". Genomics. 32 (2): 295–296. doi:10.1006/geno.1996.0120. PMID 8833160.
- Yoshimi N, Futamura T, Bergen SE, Iwayama Y, Ishima T, Sellgren C, et al. (November 2016). "Cerebrospinal fluid metabolomics identifies a key role of isocitrate dehydrogenase in bipolar disorder: evidence in support of mitochondrial dysfunction hypothesis". Molecular Psychiatry. 21 (11): 1504–1510. doi:10.1038/mp.2015.217. PMC 5078854. PMID 26782057.
- Zhou J, Yong WP, Yap CS, Vijayaraghavan A, Sinha RA, Singh BK, et al. (April 2015). "An integrative approach identified genes associated with drug response in gastric cancer". Carcinogenesis. 36 (4): 441–451. doi:10.1093/carcin/bgv014. PMID 25742747.
Further reading
- Sandoval N, Bauer D, Brenner V, Coy JF, Drescher B, Kioschis P, et al. (July 1996). "The genomic organization of a human creatine transporter (CRTR) gene located in Xq28". Genomics. 35 (2): 383–385. doi:10.1006/geno.1996.0373. PMID 8661155.
- Kim YO, Koh HJ, Kim SH, Jo SH, Huh JW, Jeong KS, et al. (December 1999). "Identification and functional characterization of a novel, tissue-specific NAD(+)-dependent isocitrate dehydrogenase beta subunit isoform". The Journal of Biological Chemistry. 274 (52): 36866–36875. doi:10.1074/jbc.274.52.36866. PMID 10601238.
- Weiss C, Zeng Y, Huang J, Sobocka MB, Rushbrook JI (February 2000). "Bovine NAD+-dependent isocitrate dehydrogenase: alternative splicing and tissue-dependent expression of subunit 1". Biochemistry. 39 (7): 1807–1816. doi:10.1021/bi991691i. PMID 10677231.
- Hartley JL, Temple GF, Brasch MA (November 2000). "DNA cloning using in vitro site-specific recombination". Genome Research. 10 (11): 1788–1795. doi:10.1101/gr.143000. PMC 310948. PMID 11076863.
- Simpson JC, Wellenreuther R, Poustka A, Pepperkok R, Wiemann S (September 2000). "Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing". EMBO Reports. 1 (3): 287–292. doi:10.1093/embo-reports/kvd058. PMC 1083732. PMID 11256614.
- Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, et al. (December 2002). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proceedings of the National Academy of Sciences of the United States of America. 99 (26): 16899–16903. Bibcode:2002PNAS...9916899M. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Gerhard DS, Wagner L, Feingold EA, Shenmen CM, Grouse LH, Schuler G, et al. (October 2004). "The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC)". Genome Research. 14 (10B): 2121–2127. doi:10.1101/gr.2596504. PMC 528928. PMID 15489334.
- Wiemann S, Arlt D, Huber W, Wellenreuther R, Schleeger S, Mehrle A, et al. (October 2004). "From ORFeome to biology: a functional genomics pipeline". Genome Research. 14 (10B): 2136–2144. doi:10.1101/gr.2576704. PMC 528930. PMID 15489336.
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, et al. (October 2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–1178. Bibcode:2005Natur.437.1173R. doi:10.1038/nature04209. PMID 16189514. S2CID 4427026.
- Mehrle A, Rosenfelder H, Schupp I, del Val C, Arlt D, Hahne F, et al. (January 2006). "The LIFEdb database in 2006". Nucleic Acids Research. 34 (Database issue): D415 – D418. doi:10.1093/nar/gkj139. PMC 1347501. PMID 16381901.
- Soundar S, O'Hagan M, Fomulu KS, Colman RF (July 2006). "Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent isocitrate dehydrogenase". The Journal of Biological Chemistry. 281 (30): 21073–21081. doi:10.1074/jbc.M602956200. PMID 16737955.
- Bzymek KP, Colman RF (May 2007). "Role of alpha-Asp181, beta-Asp192, and gamma-Asp190 in the distinctive subunits of human NAD-specific isocitrate dehydrogenase". Biochemistry. 46 (18): 5391–5397. doi:10.1021/bi700061t. PMID 17432878.
| Enzymes | |
|---|---|
| Activity | |
| Regulation | |
| Classification | |
| Kinetics | |
| Types |
|
Categories: