| Revision as of 04:16, 2 February 2007 editSeans Potato Business (talk | contribs)Extended confirmed users5,759 editsNo edit summary← Previous edit | Revision as of 04:23, 2 February 2007 edit undoLuna Santin (talk | contribs)65,325 edits closing ref tags -- <ref>stuff</ref>Next edit → | ||
| Line 1: | Line 1: | ||
| ]s of ] GAL4 are fused to two proteins being tested for interactions. The two proteins are termed "bait" and "library". If "bait" and "library" interact, transcription of "reporter gene" occurs. Otherwise, "reporter gene" is silent.]] | ]s of ] GAL4 are fused to two proteins being tested for interactions. The two proteins are termed "bait" and "library". If "bait" and "library" interact, transcription of "reporter gene" occurs. Otherwise, "reporter gene" is silent.]] | ||
| '''Two-hybrid screening''' is a ] technique used to discover ] (Young, 1998) and ] <ref name="joung" |
'''Two-hybrid screening''' is a ] technique used to discover ] (Young, 1998) and ] <ref name="joung">{{cite journal | ||
| |author=Joung J, Ramm E, Pabo C | |author=Joung J, Ramm E, Pabo C | ||
| |title=A bacterial two-hybrid selection system for studying protein-DNA and protein-protein interactions | |title=A bacterial two-hybrid selection system for studying protein-DNA and protein-protein interactions | ||
| Line 17: | Line 17: | ||
| == History == | == History == | ||
| Pioneered by Fields and Song in 1989, the technique was originally designed to detect protein-protein interactions using the GAL4 transcriptional activator of the yeast '']''. The GAL4 protein activated transcription of a protein involved in galactose utilization which formed the basis of selection (Fields and Song, 1989). Since then, the same principle has been adapted to describe many alternative methods including some that detect protein-DNA interactions and use '']'' instead of yeast <ref name="hurt" |
Pioneered by Fields and Song in 1989, the technique was originally designed to detect protein-protein interactions using the GAL4 transcriptional activator of the yeast '']''. The GAL4 protein activated transcription of a protein involved in galactose utilization which formed the basis of selection (Fields and Song, 1989). Since then, the same principle has been adapted to describe many alternative methods including some that detect protein-DNA interactions and use '']'' instead of yeast <ref name="hurt">{{cite journal | ||
| |author=Hurt J, Thibodeau S, Hirsh A, Pabo C, Joung J | |author=Hurt J, Thibodeau S, Hirsh A, Pabo C, Joung J | ||
| |title=Highly specific zinc finger proteins obtained by directed domain shuffling and cell-based selection | |title=Highly specific zinc finger proteins obtained by directed domain shuffling and cell-based selection | ||
Revision as of 04:23, 2 February 2007
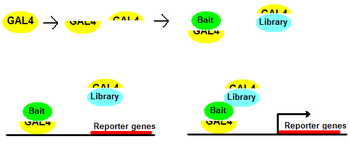
Two-hybrid screening is a molecular biology technique used to discover protein-protein interactions (Young, 1998) and protein-DNA interactions by testing for physical interactions (such as binding) between two proteins or a single protein and a DNA molecule, respectively. One protein is termed the bait and the other is a prey or library.
The premise behind the test is the activation of downstream reporter gene(s) by the binding of a transcription factor onto an upstream activating sequence (UAS). For the purposes of two-hybrid screening, the transcription factor is split into two separate fragments, called the Binding Domain (BD) and Activating Domain (AD). The BD is the domain responsible for binding to the UAS and the AD is the domain responsible for activation of transcription (Joung et al., 2000; Young, 1998).
History
Pioneered by Fields and Song in 1989, the technique was originally designed to detect protein-protein interactions using the GAL4 transcriptional activator of the yeast Saccharomyces cerevisiae. The GAL4 protein activated transcription of a protein involved in galactose utilization which formed the basis of selection (Fields and Song, 1989). Since then, the same principle has been adapted to describe many alternative methods including some that detect protein-DNA interactions and use Escherichia coli instead of yeast (Joung et al., 2000; Young, 1998).
Basic Premise
Key to the two-hybrid screen, is that in most eukaryotic transcription factors, the activating and binding domains are modular and can function in close proximity to each other without direct binding. This means that even though the transcription factor is split into two fragments, it can still activate transcription when the two fragments are indirectly connected.
The most common screening approach is the yeast two-hybrid assay. This system often utilizes a genetically engineered strain of yeast in which the biosynthesis of certain nutrients (usually amino acids or nucleic acids) is lacking. When grown on media that lacks these nutrients, the yeast fail to survive. This mutant yeast strain can be made to incorporate foreign DNA in the form of plasmids. In yeast two-hybrid screening, separate bait and prey plasmids are simultaneously introduced into the mutant yeast strain.
Plasmids are engineered to produce a protein product in which the DNA-binding domain (BD) fragment is fused onto a protein while another plasmid is engineered to produce a protein product in which the activation domain (AD) fragment is fused onto another protein. The protein fused to the BD may be referred to as the bait protein and is typically a known protein that the investigator is using to identify new binding partners. The protein fused to the AD may be referred to as the the prey protein can be either a single known protein or a library of known or unknnown proteins. In this context, a library may consist of a collection of protein-encoding sequences that represent all the proteins expressed in a particular organism or tissue or may be generated by synthesising random DNA sequences (Hurt et al., 2003). Regardless of the source, they are subsequently incoorporated into the protein-encoding sequence of a plasmid which are then transfected into the cells chosen for the screening method (Hurt et al., 2003). This technique, when using a library, assumes that each cell is transfected with no more than a single plasmid and that therfore, each cell ultimately expresses no more than a single member from the protein library.
If the bait and prey proteins interact (i.e. bind), then the AD and BD of the transcription factor are indirectly connected, bringing the AD domain in proximity to the transcription start site and transcription of reporter gene(s) can occur. If the two proteins do not interact, there is no transcription of the reporter gene. In this way, a successful interaction between the fused protein is linked to a change in the cell phenotype (Young, 1998).
The challenge of separating those cells which express proteins which happen to interact with their counterpart fusion proteins, from those which are not, is addressed in the following section, 'Reporter and selection genes'.
Reporter and selection genes
In order to link the interaction to a change in observable phenotype, a reporter gene is provided with the upstream activation sequence (UAS) to which the binding domain binds, resulting in gene expression in successful cases of interaction. Since its inception in 1989, the technique has been combined with a number of different reporter genes which can allow selection through a simple colour change or through automatic death of cells in which the interaction does or does not take place (Young, 1998).
The lacZ reporter will allow the highlighting of cells in which the UAS-BD-AD interaction is taking place (Young, 1998). β-galactosidase, the gene product of lacZ produces a blue colouration through the metabolism of X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactoside) which allows the experimenter to manually choose the individuals which host proteins displaying the required interaction (Joung et al, 2000).
Manual differentiation of cells according to colour may be acceptable for small numbers of cells, as when investigating a small number of proteins, but when a large library of proteins must be screened, some automation is necessary. This automation is provided by a number of genes ad gene systems which either cause the death of cells that aren't hosting interactions or vice cersa, leaving only the cells expressing the proteins of interest.
For more autonomy, alternative reporter genes exist that enable automatic deletion of individuals that fail to express the reporter or vice versa.
Positive selection
In positive selection, the individuals that host a successful interaction are able to live on the provided media, while those failing to host this interaction die. This can be achieved by combining a media that is lacking an essential nutrient with an organism dependent on expression of the reporter gene in order to produce this nutrient. Examples include the HIS3 gene, encoding a protein required for histidine synthesis, the LEU2 gene, encoding a protein required for leucine synthesis and the URA3 gene, encoding a protein required for uracil synthesis (Young, 1998).
The positive selection method is used in investigations in which the investigators are aiming to discover proteins which interact. Applications include the descovery of homologues in other species and the discovery of members of a protein family (containing a particular domain and therefore binding to a compatible domain) (Young, 1998).
Counter-selection
Alternatively, reporter genes conferring sensitivity to an agent supplied in the media, will kill cells expressing the reporter, thereby removing cells that host an interaction. Examples of genes used in the counter selection method include CYH2 (confers sensitivity to cycloheximide), CAN1 and URA3 (Young, 1998). Alternatively, using repressor domains or other negative regulators in place of activation domains, counter-selection may be achieved using the same reporter genes used in positive selection methods. One such negative regulator is the yeast GAL80 domain which binds and inactivates the transcriptional activator region of GAL4 (Young, 1998).
Such counter-selection methods may be used in investigations aiming to discover a mutation, chemical or protein that will interfere with the interaction.
Controlling sensitivity
The Escherichia coli-derived Tet-R repressor can be used in line with a conventional reporter gene and can be controlled by tetracycline or doxicycline (Tet-R inhibitors). Thus the expression of Tet-R is controlled by the standard two-hybrid system but the Tet-R in turn controls (represses) the expression of a previously mentioned reporter such as HIS3, through its Tet-R promoter. Tetracycline or its derivatives can then be used to regulate the sensitivity of a system utilising Tet-R (Young, 1998).
Sensitivity may also be controlled by varying the dependency of the cells on their reporter genes. For example, this is effected by altering the concentration of histidine in the growth medium for his3-dependent cells and altering the concentration of streptomycin for aadA dependent cells (Hurt et al., 2003; Joung et al., 2000).
Some projects may utilise a dual reporter system in which two reporter genes are used in order to help substantiate the interaction (Hurt et al., 2003; Young, 1998).
Non-fusion proteins
A third, non-fusion protein may be co-expressed with the two fusion proteins. Depending on the investigation, the third protein may modify one of the fusion proteins or mediate or interfere with their interaction (Young, 1998).
Coexpression of the third protein may be necessary for modification or activation of one or both of the fusion proteins. For example S. cerevisiae possesses no endogenous tyrosine kinase. If an investigation involves a protein that requires tyrosine phosphorylation, the kinase must be supplied in the form of a tyrosine kinase gene (Young, 1998).
The non-fusion protein may mediate the interaction by binding both fusion proteins simultaneously, as in the case of ligand-dependent receptor dimerisation (Young, 1998).
For a protein with an interacting partner, its functional homology to other proteins may be assessed by supplying the third protein in non-fusion form which then may or may not compete with the fusion-protein for its binding partner. Binding between the third protein and the other fusion protein will interupt the formation of the reporter expression activation complex and thus reduce reporter expression, leading to the distinguishing change in phenotype (Young, 1998).
One-hybrid, three-hybrid and one-two-hybrid variants
One-hybrid
The one-hybrid variation of this technique is designed to investigate protein-DNA interactions and uses a single fusion protein in which the AD is linked directly to the binding domain. The binding domain in this case however is not necessarily of fixed sequence as in two-hybrid analysis but may be constituted by a library. This library is selected against the desired target sequence which is inserted in the promoter region of the reporter gene construct. In a positive-selection system, a binding domain which successfully binds the UAS and allows transcription will thus be selected (Young, 1998).
Three-hybrid
RNA-protein interactions have been investigated through a three-hybrid variation of the two-hybrid technique. In this case, a hybrid RNA molecule serves to adjoin together the two protein fusion domains which aren't intended to interact with each other but rather the intermidiary RNA molecule (through their RNA-binding domains) (Young, 1998). Techniques involving non-fusion proteins that perform a similar function, as described in the 'non-fusion proteins' section above, may also be referred to as three-hybrid methods.
One-two-hybrid
Simultaneous use of the one- and two-hybrid methods (that is, simultaneous protein-protein and protein-DNA interaction) is known as a one-two-hybrid approach and expected to increase the stringency of the screen (Young, 1998).
Applications
Determination of sequences crucial to interaction
By mutating specific DNA base-pairs, the importance of the corresponding amino acids, in maintaining the interaction, can be determined (Young, 1998).
Drug and poison discovery
Protein-protein signalling interactions pose suitable therapeutic targets due to their specificity and pervasiveness. The random drug discovery approach utilises compound banks that comprise random chemical structures and demands a high-throughput method in order to test these structures in their intended target (Young, 1998).
The cell chosen for the investigation can be specifically engineered to mirror the molecular aspect that the investigator intends to study and then used to identify new human or animal therapeutics or anti-pest agents (Young, 1998).
Determination of protein function
By determination of the interaction partners of unknown proteins, the possible functions of these new proteins may be inferred (Young, 1998). This can be done using a single known protein against a library of unknown proteins or conversely, by selecting from a library of known proteins using a single protein of unknown function (Young, 1998).
Zinc finger protein selection
To select zinc finger proteins (ZFPs) for protein engineering, methods adapted from the two-hybrid screening technique have been used with success (Hurt et. al., 2003; Joung et. al., 2000). A ZFP is itself a DNA-binding protein used in the construction of custom DNA-binding domains that bind to a desired DNA sequence .
By using a selection gene with the desired target sequence included in the UAS and randomising the relevant amino acid sequences to produce a ZFP library, cells that host a DNA-ZFP interaction with the required characteristics can be selected. Each ZFP typically recognises only 3-4 base pairs, so to prevent recognition of sites outside the UAS, the randomised ZFP is engineered into a 'scaffold' consisting of another two ZFPs of constant sequence. The UAS is thus designed to include the target sequence of the constant scaffold in addition to the sequence for which a ZFP is being selected (Hurt et. al., 2003; Joung et. al., 2000).
Host organism
Although theoretically, any living cell might be used as the background to a two-hybrid analysis, there are practical considerations that dictate which will be chosen. The chosen cell line should be relatively cheap and easy to culture and sufficiently robust to withstand application of the investigative methods and reagents (Young, 1998).
Yeast
S. cerevisiae was the model organism used during the two-hybrid technique's inception. It has several characteristics that make it a robust organism to host the interaction, including the ability to form tertiary protein structures, neutral internal pH, enhanced ability to form disulphide bonds and reduced-state glutathione among other cytosolic buffer factors, to maintain a hospitable internal environment (Young, 1998). The yeast model can be manipulated through non-molecular techniques and its complete gene sequence is known (Young, 1998). Yeast systems are tolerant of diverse culture conditions and harsh chemicals which could not be applied to mammalian tissue cultures (Young, 1998).
E. coli
Strengths and weaknesses
Two-hybrid screens are now routinely performed in many labs. They can provide an important first hint for the identification of interaction partners. Moreover, the assay is scalable which makes it possible to screen for interactions among many proteins.
The main critisism applied to the yeast two-hybrid screen of protein-protein interactions is the possibility of a high number of false positive (and false negative) identifications. The exact rate of false positive results is not known, but estimates are as high as 50% . The reason for this high error rate lies in the principle of the screen: The assay investigates the interaction between (i) overexpressed (ii) fusion proteins in the (iii) yeast (iv) nucleus. Each of these points (i-iv) alone can give rise to false results. For example, overexpression can result in non-specific interactions. Moreover, a mammalian protein is sometimes not correctly modified in yeast (e.g. missing phosphorylation), which can also lead to false results. Finally, some proteins might specifically interact when they are co-expressed in the yeast, although in reality they are never present in the same cell at the same time. Due to the combined effects of all error sources the overall confidence of the yeast two-hybrid assay is rather low. This means that all interactions have to be confirmed by a high confidence assay like co-immunoprecipitation of the endogenous proteins. However, this is a difficult task, especially for large scale protein-protein interaction data.
See also
- Phage display, an alternative method for detecting protein-protein and protein-DNA interactions
References
- Joung J, Ramm E, Pabo C (2000). "A bacterial two-hybrid selection system for studying protein-DNA and protein-protein interactions". Proc Natl Acad Sci U S A. 97 (13): 7382–7. PMID 10852947.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Hurt J, Thibodeau S, Hirsh A, Pabo C, Joung J (2003). "Highly specific zinc finger proteins obtained by directed domain shuffling and cell-based selection". Proc Natl Acad Sci U S A. 100 (21): 12271–6. PMID 14527993.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Gommans W, Haisma H, Rots M (2005). "Engineering zinc finger protein transcription factors: the therapeutic relevance of switching endogenous gene expression on or off at command". J Mol Biol. 354 (3): 507–19. PMID 16253273.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Deane C, Salwiński Ł, Xenarios I, Eisenberg D (2002). "Protein interactions: two methods for assessment of the reliability of high throughput observations". Mol Cell Proteomics. 1 (5): 349–56. PMID 12118076.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
- Deane CM, Salwinski L, Xenarios I, Eisenberg D. (2002) Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol Cell Proteomics. 1:349-56. PMID: 12118076
- Gommans, W. M., H. J. Haisma and M. G. Rots (2005). "Engineering zinc finger protein transcription factors: the therapeutic relevance of switching endogenous gene expression on or off at command." Journal of Molecular Biology 354(3): 507-19.
- Hurt, J. A., S. A. Thibodeau, A. S. Hirsh, C. O. Pabo and J. K. Joung (2003). "Highly specific zinc finger proteins obtained by directed domain shuffling and cell-based selection." Proc Natl Acad Sci U S A 100(21): 12271-6.
- Joung, J. K., E. I. Ramm and C. O. Pabo (2000). "A bacterial two-hybrid selection system for studying protein-DNA and protein-protein interactions." Proc Natl Acad Sci U S A 97(13): 7382-7.
- Young, K. H. (1998). "Yeast two-hybrid: so many interactions, (in) so little time." Biol Reprod 58(2): 302-11.
External links
- Detail on sister technique two-hybrid system
- Science Creative Quarterly's overview of the yeast two hybrid system