| Revision as of 11:47, 13 May 2009 editSMcCandlish (talk | contribs)Autopatrolled, Extended confirmed users, Page movers, File movers, New page reviewers, Pending changes reviewers, Rollbackers, Template editors201,792 editsm verb tense← Previous edit | Revision as of 15:15, 13 May 2009 edit undoWapondaponda (talk | contribs)2,285 edits →Recent Medieval North African/Near Eastern admixture: imageNext edit → | ||
| Line 149: | Line 149: | ||
| === Recent Medieval North African/Near Eastern admixture=== | === Recent Medieval North African/Near Eastern admixture=== | ||
| ] | |||
| A 2004 study showed that E1b1b1b is found present, albeit at low levels throughout Southern Europe (ranging from 1.5% in Northern Italians, 2.2% in Central Italians, 1.6% in southern Spaniards, 3.5% in the French, 4% in the Northern Portuguese, 12.2% in the southern Portuguese and 41.2% in the genetic isolate of the Pasiegos from Cantabria)<ref></ref>. The findings of this latter study contradict a more thorough analysis Y-chromosome analysis of the Iberian peninsula according to which haplogroup E1b1b1b surpasses frequencies of 10% in Southern Spain. The study points only to a very limited influence from northern Africa and the Middle East both in historic and prehistoric times.<ref></ref> The absence of microsatellite variation suggests a very recent arrival from North Africa consistent with historical exchanges across the Mediterranean during the period of Islamic expansion, namely of ] populations.<ref name="Semino">* Semino et al. (2004) </ref>. A study restricted to ], concerning Y-chromosome lineages, revealed that "The mtDNA and Y data indicate that the Berber presence in that region dates prior to the Moorish expansion in 711 AD... Our data indicate that male Berbers, unlike sub-Saharan immigrants, constituted a long-lasting and continuous community in the country".<ref></ref> | A 2004 study showed that E1b1b1b is found present, albeit at low levels throughout Southern Europe (ranging from 1.5% in Northern Italians, 2.2% in Central Italians, 1.6% in southern Spaniards, 3.5% in the French, 4% in the Northern Portuguese, 12.2% in the southern Portuguese and 41.2% in the genetic isolate of the Pasiegos from Cantabria)<ref></ref>. The findings of this latter study contradict a more thorough analysis Y-chromosome analysis of the Iberian peninsula according to which haplogroup E1b1b1b surpasses frequencies of 10% in Southern Spain. The study points only to a very limited influence from northern Africa and the Middle East both in historic and prehistoric times.<ref></ref> The absence of microsatellite variation suggests a very recent arrival from North Africa consistent with historical exchanges across the Mediterranean during the period of Islamic expansion, namely of ] populations.<ref name="Semino">* Semino et al. (2004) </ref>. A study restricted to ], concerning Y-chromosome lineages, revealed that "The mtDNA and Y data indicate that the Berber presence in that region dates prior to the Moorish expansion in 711 AD... Our data indicate that male Berbers, unlike sub-Saharan immigrants, constituted a long-lasting and continuous community in the country".<ref></ref> | ||
Revision as of 15:15, 13 May 2009
| This article needs attention from an expert in Molecular and Cellular Biology. Please add a reason or a talk parameter to this template to explain the issue with the article. WikiProject Molecular and Cellular Biology may be able to help recruit an expert. (November 2008) |
| This article's factual accuracy is disputed. Relevant discussion may be found on the talk page. Please help to ensure that disputed statements are reliably sourced. (July 2007) (Learn how and when to remove this message) |
The genetic history of Europe can be inferred by observing the patterns of genetic diversity across the continent and comparing them with the patterns on the adjacent land masses. These patterns can be found by using classical genetic markers or by using molecular genetics (autosomal, Y-chromosome and mitochondrial DNA). Most data is from modern populations, but there is a small amount of information from ancient DNA. European populations have a complicated demographic and genetic history, including many layers of successive migrations between different time periods, from the first appearance of Homo sapiens in the Upper Paleolithic to contemporary immigration.

Relation of Neanderthals and modern humans
Before modern humans arrived in Europe some 45,000 years ago, the continent was inhabited by Neanderthals, who did not die out until about 25,000 years ago. There continues to be much speculation as to whether the two genetic groups interbred. Analysis of the DNA distributions of the two groups shows significant difference, so any such offspring were unlikely to be fertile.
Relation of present population to other populations
Classical genetics studies
| Africa | Oceania | East Asia | Europe | |
|---|---|---|---|---|
| Oceania | 24.7 | |||
| East Asia | 20.6 | 10 | ||
| Europe | 16.6 | 13.5 | 9.7 | |
| America | 22.6 | 14.6 | 8.9 | 9.5 |
In the 1990s, a study by Luigi Luca Cavalli-Sforza of the Stanford University School of Medicine, using 120 blood polymorphisms, provided information on genetic relatedness of the various continental populations. Genetic distance is a measure of genetic difference between two populations. It is based on the principle that two populations that share similar frequencies of a trait are more closely related than populations that have more divergent frequencies of the trait. In its simplest form, it is the difference in frequencies of a particular trait between two populations. For example, the frequency of RH negative individuals is 50.4% among Basques, 41.2% in France and 41.1% in England. Thus regarding the RH negative trait, the genetic difference between the Basques and French is 9.2% and the genetic difference between the French and the English is 0.1%. Averaged over several traits this approach can give a measure of the overall genetic relatedness of various populations.
According to the study, all non-African populations are more closely related to each other than to Africans; this supports the hypothesis that all non-Africans descend from a single African population. The genetic distance from Africa to Europe (16.6) is shorter than the genetic distance from Africa to East Asia (20.6), and even much shorter than that from Africa to Australia (24.7). The simplest explanation for this short genetic distance, according to Cavalli-Sforza, is that substantial gene exchange has taken place between the nearby continents. Cavalli-Sforza suggests that this admixture took place 30,000 years ago. The overall contributions from Asia and Africa were estimated to be around two-thirds and one-third, respectively. Europe's genetic variation is about a third of that of other continents.
According to Guglielmino et al. (1990),
Principal coordinate analysis shows that Lapps/Sami are almost exactly intermediate between people located geographically near the Ural mountains and speaking Uralic languages, and central and northern Europeans. Hungarians and Finns are definitely closer to Europeans. An analysis of genetic admixture between Uralic and European ancestors shows that Lapps/Sami are slightly more than 50% European, Hungarians are 87% European, and Finns are 90% European. There is basic agreement between these conclusions and historical data on Hungary. Less is known about Finns and very little about Lapps/Sami.
DNA studies
Studies of the genetic history of Europe have been done using mitochondrial DNA (mtDNA), Y chromosome DNA and autosomal DNA. The first allows to trace female lines, the second allows to trace male lines, while the third provides many possible markers (over 500,0000 have been used) but allows descents only to be determined on a statistical basis because of recombination of male/female DNA. By comparing the polymorphisms found in DNA analyses, haplogroup trees have been defined. These have made use of both single nucleotide polymorphisms and short tandem repeats, which produce information about long and short timescale changes respectively.
Worldwide DNA studies have shown that a group of Homo sapiens left Africa for the Yemen some 80,000 years ago. Some of their descendants entered Europe about 30,000 years later. The Y-chromosome and the mtDNA haplogroups found in Europe differ in their frequency distributions from those in Africa, Asia and the Middle East. However some predominantly European haplogroups are found in Asia and vice versa. A similar correspondence is found between North Africa and Europe.
This relationship of European to Asian populations has been shown for example by the spread of the Y-chromosome marker Haplogroup N, in Northern Europe. . Several studies strongly suggest a pattern of migrations from Asia to Northern Europe over the last 4000 years. For example, the studies by Zerjal et al. 1997, Su et al. 1999, and Lell et al. 2002 established a significant presence of this Asian marker in different European peoples, ranging from a 52% in Finns, 47% in Lithuanians, 37% in Estonians and 32% in Latvians to 14% in Russians, 11% in Ukranians, 8% in North Swedes, 6% in Gotlanders, 6% in Norwegians, 4% in Poles, 3% in Germans and 1% in Turks, among others.
European population structure
| The neutrality of this section is disputed. Relevant discussion may be found on the talk page. Please do not remove this message until conditions to do so are met. (December 2008) (Learn how and when to remove this message) |
In 2006, an autosomal analysis comparing samples from various European populations concluded that “there is a consistent and reproducible distinction between ‘northern’ and ‘southern’ European population groups”. Most individual participants with southern European ancestry (Italian, Greek, Armenian, Portuguese, and Spanish) have >85% membership in the ‘southern’ population; and most northern, western, eastern, and central Europeans have >90% in the ‘northern’ population group. Ashkenazi Jewish as well as Sephardic Jewish origin also showed >85% membership in the ‘southern’ population, consistent with a later Mediterranean origin of these ethnic groups". Many of the participants in this study were actually American citizens who self identified with different European ethnicities and were not Europeans.
Somewhat contradicting these findings, a similar study in 2007 using samples exclusively from Europe found that the most important genetic differentiation in Europe occurs on a line from the north to the south-east (northern Europe to the Balkans), with another east-west axis of differentiation across Europe. Its findings were consistent with earlier results based on mtDNA and Y-chromosonal DNA that support the theory that modern Iberians (Spanish and Portuguese) hold the most ancient European genetic ancestry, as well as separating Basques and Sami from other European populations. It confirmed that the English and Irish cluster with other Northern and Eastern Europeans such as Germans and Poles, while some Basque and Italian individuals also clustered with Northern Europeans. Despite these stratifications, it noted the unusually high degree of European homogeneity: "there is low apparent diversity in Europe with the entire continent-wide samples only marginally more dispersed than single population samples elsewhere in the world."
In fact, according to another European wide study, the main components in the European genomes appear to derive from ancestors whose features were similar to those of modern Basques and Near Easterners, with average values greater than 35% for both these parental populations, regardless of whether or not molecular information is taken into account. The lowest degree of both Basque and Near Eastern admixture is found in Finland, whereas the highest values are, respectively, 70% ("Basque") in Spain and more than 60% ("Near Eastern") in the Balkans.
In 2008, two international research teams published analyses of large-scale genotyping of large samples of Europeans, revealing relatively little genetic differentiation between the various European populations sampled. (The samples for these papers used overlapped somewhat, though the papers were not collaborative.) But the number of loci included in the analysis was sufficient to detect the geographic region an individual comes from to within about 840 km (for 90% of individuals), as long as the individual's recent ancestry is from that region (for example if an individual has parents from different regions of Europe, then that individual will be placed on the map exactly equidistant between the parent's populations of origin, and not near either parental population). Southern Europeans have more genetic diversity, having both less linkage disequilibrium and greater heterozygosity, indicating a larger effective population size and/or population expansion from southern to northern Europe, as expected. Populations did not form clusters as previous studies have found (see Seldin et al. 2006 and Bauchett et al. 2007), but showed a correlation between genetic distance and geographic distance. The researchers take this observation to imply that, genetically speaking, Europeans are not distributed into discrete, populations.
Western Europe substructure
It is thought that ancient Iberia served as a refuge for palaeolithic humans during the last major glaciation when environments further north were too cold and dry for continuous habitation. When the climate warmed into the present interglacial, populations would have rapidly spread north along the west European coast. Genetically, in terms of Y-chromosomes and mtDNA, inhabitants of Britain and Ireland are closely related to the Basques, reflecting their common origin in this refugial area. Basques, along with Irish, show the highest frequency of the Y-chromosome DNA haplogroup R1b in Western Europe; some 95% of native Basque men have this haplogroup. The rest is mainly I and a minimal presence of E3b. The Y-chromosome and mtDNA relationships between Basques and the people of Ireland and Wales are closer than of the Basques to neighbouring areas of Spain, where similar ethnically "Spanish" people now live in close proximity to the Basques, although this genetic relationship is also very strong among Basques and other Spaniards. In fact, as Stephen Oppenheimer has stated in The Origins of the British (2006), although Basques have been more isolated than other Iberians, they are a population representative of south western Europe. As to the genetic relationship among Basques, Iberians and Britons, he also states (pages 375 and 378):
By far the majority of male gene types in the British Isles derive from Iberia (modern Spain and Portugal), ranging from a low of 59% in Fakenham, Norfolk to highs of 96% in Llangefni, north Wales and 93% Castlerea, Ireland. On average only 30% of gene types in England derive from north-west Europe. Even without dating the earlier waves of north-west European immigration, this invalidates the Anglo-Saxon wipeout theory... ...75-95% of British and Irish (genetic) matches derive from Iberia...Ireland, coastal Wales, and central and west-coast Scotland are almost entirely made up from Iberian founders, while the rest of the non-English parts of Britain and Ireland have similarly high rates. England has rather lower rates of Iberian types with marked heterogeneity, but no English sample has less than 58% of Iberian samples...
Brian Sykes, in his book based on genetics Blood of the Isles (2006) comes to similar conclusions. Some quotations from the book follow. (Note that Sykes uses the terms "Celts" and "Picts" to designate the pre-Roman inhabitants of the Isles rather than as linguistic terms.)
he presence of large numbers of Jasmine’s Oceanic clan ... says to me that there was a very large-scale movement along the Atlantic seaboard north from Iberia, beginning as far back as the early Neolithic and perhaps even before that. The number of exact and close matches between the maternal clans of western and northern Iberia and the western half of the Isles is very impressive, much more so than the much poorer matches with continental Europe.
The genetic evidence shows that a large proportion of Irish Celts, on both the male and female side, did arrive from Iberia at or about the same time as farming reached the Isles. (...)
The connection to Spain is also there in the myth of Brutus.... This too may be the faint echo of the same origin myth as the Milesian Irish and the connection to Iberia is almost as strong in the British regions as it is in Ireland. (...)
They are from the same mixture of Iberian and European Mesolithic ancestry that forms the Pictish/Celtic substructure of the Isles.
Here again, the strongest signal is a Celtic one, in the form of the clan of Oisin, which dominates the scene all over the Isles. The predominance in every part of the Isles of the Atlantic chromosome (the most frequent in the Oisin clan), with its strong affinities to Iberia, along with other matches and the evidence from the maternal side convinces me that it is from this direction that we must look for the origin of Oisin and the great majority of our Y-chromosomes. The sea routes of the Atlantic fringe conveyed both men and women to the Isles.
Oppenheimer states that most Western Europeans are also mainly of Iberian origins, but with some input from Eastern Europe.
Eastern Europe substructure
It is thought that there were refugia in the Balkans and the Ukraine that acted as source areas for the repopulation of Europe after the LGM. Associated with these areas are Y-chromosomes I and R1a respectively. People from these refuges reached north-west Europe, while R1a also spread into central Asia and as far as Pakistan and India. There was a major incursion of Halpogroup N into north-eastern Europe. South-eastern Europe shows the incursion of several haplogroups mainly found in the Middle East.
Haplogroups in Europe
Human Y-chromosome DNA haplogroups

There are three major Y-chromosome DNA haplogroups which largely account for most of Europe's present-day population.
- Haplogroup R1b is common on the western Atlantic coast of Europe, from the Iberian Peninsula (comprising Spain and Portugal) to Ireland, Wales, England and Scotland, and Jutland.
- Haplogroup I is common across Germany, the Netherlands, Austria, and up into Scandinavia, as well as a very high amount in the western Balkans.
- Haplogroup R1a is common in Central and Eastern Europe (and is also common in Central Asia and the Indian subcontinent).
Most common of all haplogroups among western Europeans is R1b. The following values of Hg R1b are: Welsh: 89.0%; Basques: 88.1%; Irish: 81.5%; Scots: 77.1%; British: 68.8; Non-Basque Spaniards: 68.0 (Catalans: 79.2; Andalusians: 65.5); Belgians: 63.0; Portuguese (North): 62.0%; Italians (North-central): 62.0; English (Central): 61.9%; Portuguese (South): 56.0%; French: 52.2%; Danes: 41.7%, Germans: 47.9; Czechs & Slovaks: 35.6%; Italians (Calabria): 32.4; Norwegians: 25.9; Greeks: 22.8%; Italians (Sardinia): 22.1%; Slovenians: 21%; Swedes: 20.0; Romanians: 18.0%; Albanians: 17.6%; Bulgarians: 17.0%; Polish: 16.4%; Turks: 16.3%; Croatians (mainland): 15.7%; Hungarians: 13.3%; Serbs: 10.6%; Cypriots: 9.0%.
Among European populations, diversity is highest in Eastern Europe, despite lower frequencies. Diversity analysis indicates that all European variants of R1b shared an existence in Central Asia (Kazakhstan) before migrating to Russia and then splitting into two major migrations, moving primarily along rivers and coastlines.
Each haplogroup clade may have subclades. R1a and R1b are subclades of Haplogroup R (Y-DNA) Two main subgroups of Haplogroup I (Y-DNA) are I-M253/I-M307/I-P30/I-P40, which according to the International Society of Genetic Genealogy "has highest frequency in Scandinavia, Iceland, and northwest Europe" and I-S31, which according to the International Society of Genetic Genealogy, "includes I-P37.2, which is the most common form in the Balkans and Sardinia, and I-S23/I-S30/I-S32/I-S33, which reaches its highest frequency along the northwest coast of continental Europe."
The cause and extent of the changes resulting in Neolithic Europe are still debated. There is evidence both for and against a demic diffusion from the Near East: G Barbujani and L Chikhi (2006) state, "Genetic studies have failed to settle the controversy so far, because they have been interpreted in different ways... A rather heated debate followed, and is still continuing." But a recent paper by Cruciani discounts the old hypothesis that there was a significant inflow of genes from Anatolia to Europe, by way of the Balkans, during the Neolithic. Cruciani concluded that Haplogroup E3b in particular was introduced from Western Asia "any time after 17.0 ky ago". It then spread throughout Europe much later (circa 5.3 kYa) via an in situ population expansion originating in the Balkans, corresponding to the onset of the Balkan Bronze Age
Also, around 4,500 years ago, Haplogroup N3 began moving westward from west of the Ural mountains, and seems to follow closely the spread of the Finno-Ugric languages. It is also present at high frequencies in northern Russians, reflecting the absorption of Finno-Ugric tribes.
Human mitochondrial DNA haplogroups
There have been a number of studies about the mitochondrial DNA haplogroups (mtDNA) in Europe. According to the University of Oulu Library in Finland:
Classical polymorphic markers (i.e. blood groups, protein electromorphs and HLA antigenes) have suggested that Europe is a genetically homogeneous continent with a few outliers such as the Saami, Sardinians, Icelanders and Basques (Cavalli-Sforza et al. 1993, Piazza 1993). The analysis of mtDNA sequences has also shown a high degree of homogeneity among European populations, and the genetic distances have been found to be much smaller than between populations on other continents, especially Africa (Comas et al. 1997).
The mtDNA haplogroups of Europeans are surveyed by using a combination of data from RFLP analysis of the coding region and sequencing of the hypervariable segment I. About 99% of European mtDNAs fall into one of ten haplogroups: H, I, J, K, M, T, U, V, W or X (Torroni et al. 1996a). Each of these is defined by certain relatively ancient and stable polymorphic sites located in the coding region (Torroni et al. 1996a)... Haplogroup H, which is defined by the absence of a AluI site at bp 7025, is the most prevalent, comprising half of all Europeans (Torroni et al. 1996a, Richards et al. 1998)... Six of the European haplogroups (H, I, J, K, T and W) are essentially confined to European populations (Torroni et al. 1994, 1996a), and probably originated after the ancestral Caucasoids became genetically separated from the ancestors of the modern Africans and Asians.
Migrations
The prehistory of the European peoples can be traced by the examination of archaeological sites, linguistic studies, and by the examination of the DNA of the people who live in Europe now, or from recovered ancient DNA. Much of this research is ongoing, and discoveries are still being continually made, so theories rise and fall. Although it is possible to track the various migrations of people across Europe using founder analysis of DNA, most information on these movements comes from archeology.
Paleolithic and Mesolithic migrations
Further information: Paleolithic Europe and Mesolithic Europe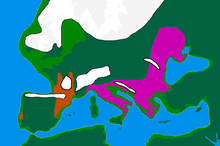
Modern humans (Cro Magnon) began to colonize Europe in the Paleolithic about 40,000 years ago, as evidenced by the spread of the Aurignacian culture. Modern humans may have arrived along two major routes either side of the Black Sea. By about 25,000 years ago, the prior inhabitants (our cousin species H. neanderthalensis) were either killed off or absorbed into the population and ultimately became extinct. Martin Richards et al. found that 21% of extant studied the mtDNA lines of Europe came from pre-glacial migrations. The two main haplogroups were U and HV, which arrived around 50,000 and 35,000 years ago respectively. Early on, HV split into Pre-V (around 26,000 years old) and the larger branch H, both of which spread over all Europe. Halpogroup H accounts for about half the gene lines in Europe, with many subgroups. The large Y-chromosome haplogroup R1 moved into Central Europe from the east 30,000 years ago and to the south-west before the LGM.
About 25,000 years ago began the last very cold period (the Last Glacial Maximum, LGM), rendering much of Europe uninhabitable. Humans may only have occupied certain regions of Europe at this time. These areas are often called refuges (or refugia) and were located along the northern Mediterranean and Black Sea coasts, as well as in the Balkans and Ukraine. As the glaciers receded from about 16,000 years ago, the populations that had occupied the refuges are thought to have begun to spread and colonise northern Europe. Pre-V's daughter group V (from about 16,300 years ago) gave the first indication of spread from the Galacian refuge, being found mostly in western and northern regions. It is found with highest frequency in Scandinavia and Lapland, but also in south-west France and with declining frequency up the Atlantic coast. Subgroups of haplogroup H seem to have originated in the Iberian refuge and spread, for example, into the British Isles. Presumably as a result of genetic drift, only one Y-chromosome subgroup (R1b) emerged from the Iberian refuge and colonised western Europe. Its frequency of occurrence declines from 90% in Basques to 50% in Germans and 0% around the eastern Baltic coast. In the north-west R1b is partly replaced by R1a, coming from an eastern refuge. There is also an exclusively European haplogroup I, deriving from the Balkans but with high frequencies in Scandinavia and northern Germany. Martin Richards estimates that overall around 50% of mtDNA arrived in the Late Upper Paleolithic, while only around 20% of Y-chromosomes did.
After a less severe cold event around 12000-10000 years ago, there was an increasing use of microliths and reliance on the coast and sea. Styles of tool making varied by location, suggesting that the population of Europe was settling down. Northern Europe was first settled in the Mesolithic. Martin Richards showed that about 11% of modern mtDNA arrived from the Middle East during the Mesolithic. These types include T, T2 and K which show a significant decline from SE to NW Europe. However Stephen Oppenheimer says that there was further gene flow from Iberia to NW Europe. He has identified four mtDNA subgroups that expanded into western Europe and nine Y-chromosome R1b descendant clusters that expanded in the Mesolithic. In northern Europe there was also mtDNA input from Asia, while the male gene flow was largely from SE Europe and Asia, including descendants of haplogroups R1a and N3, though there was also R1b input.
Neolithic migrations
Further information: Neolithic Europe and Neolithic RevolutionThe Neolithic saw the introduction of many innovations related to farming. The duration of the Neolithic varied from place to place, starting with the introduction of farming and ending with the introduction of bronze implements. In SE Europe it was approximately 7000-3000 BC while in NW Europe 4500-1700 BC. Besides the introduction of new plants and animals, the Neolithic also saw the beginning of the use of pottery. Pottery remains allow the tracing of the movement of ideas and possibly people across Europe. The period possibly also saw the spread of Indo-European languages across Europe. One hypothesis is that they spread with farming, while another is they came later from the Pontic steppes by expansion of the Kurgan people. Some academics theorise that farming was introduced by people who migrated from the Near East, and that these farmers introduced the Indo-European languages to Europe. This theory is typically associated with the Anatolian hypothesis of Indo-European origins, though it has also been argued that widespread migration is not necessary to support the theory.
Martin Richards estimated that 11% of European mtDNA is due to immigration in this period. Gene flow from SE to NW Europe seems to have continued in the Neolithic, the percentage significantly declining towards the British Isles. Classical genetics also suggested that the largest admixture to the European Paleolithic/Mesolithic stock was due to the Neolithic revolution of the 7th to 5th millennia BC. Three main mtDNA gene groups have been identified as contributing Neolithic entrants into Europe: J, T1 and U3 (in that order of importance). With others they amount up to around 20% of the gene pool. There is little published information on male immigration during the Neolithic, but Oppenheimer suggests that haplotype J1 migrated up the Danube valley while haplotypes J2 and Eb3 migrated westward along the coast of Europe at this time.
The general parent Y-chromosome Haplogroup E1b1b (formerly known as E3b), originating either in the Horn of Africa or the Near East, is by far the most common clade in North and Northeast Africa, and is also common throughout the majority of Europe, particularly in the Mediterranean and South Eastern Europe. E1b1b reaches its highest concentration in Greece and the Balkan region, but also enjoys a significant presence in other regions such as Hungary, Italy, France, Iberia and Austria. ..
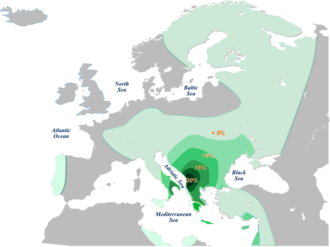
Outside of North and Northeast Africa, E1b1b's two most prevalent clades are E1b1b1a (E-M78, formerly E3b1a) and E1b1b1b (E-M81, formerly E3b1b).
E1b1b1a (also known as V-13) is the most common subclade of E1b1b and is present throughout Europe. It was originally thought to have been a marker of Neolithic migrations (perhaps coinciding with the introduction of Agriculture into Europe) from Anatolia to Europe, via the Balkans, where it enjoys the highest frequency. However, Cruciani's latest study suggests that it actually arrived into the Balkans from Western Asia during the Palaeolithic, and then spread throughout Europe much later (circa 5300 years ago) due to a population expansion originiating from within the Balkans.
Bronze and Iron Age migrations
Further information: Bronze Age Europe and Iron Age EuropeThe Bronze Age saw the development of long-distance trading networks, particularly along the Atlantic Coast and in the Danube valley. There was migration from Norway to Orkney and Shetland in this period (and to a lesser extent to mainland Scotland and Ireland). There was also migration from Germany to eastern England. Martin Richards estimated that there was about 4% mtDNA immigration to Europe in the Bronze Age. Oppenheimer could find no genetic evidence for any Iron Age migration to Britain.
One theory about the origin of the Indo-European language centres around a hypothetical Proto-Indo-European people, who are traced, in the Kurgan hypothesis, to somewhere north of the Black Sea at about 4500 BCE. They domesticated the horse, and are considered to have spread their culture and genes across Europe. It has been difficult to identify what these "Kurgan" genes might be, though the Y haplogroup R1a is a proposed marker which would indicate that the physical expansion halted in Germany and only the Kurgan culture and language went further. Another hypothesis — the Anatolian hypothesis — suggests an origin in Anatolia with a later expansion from eastern Europe.
To what extent Indo-European migrations replaced the indigenous Mesolithic peoples is debated, but a consensus has been reached that technology and language transfer played a more important role in this process than actual gene-flow.
During the Iron Age, Celts are recorded as having moved from Gaul into northern Italy, Eastern Europe and Anatolia. The relationship between the Celts of Gaul and Spain is unclear as any migration occurred before records exist.
historic non-European admixture
Roman Period admixture
During the period of the Roman Empire, historical sources show that there were many movements of people around Europe, both within and outside the Empire. These included army personnel and administrators as well as private citizens. However, compared with the total population, these movements seem mostly to have been small. No genetic information on these migrations appears to exist other than a person with a rare Yorkshire surname of African ancestry (Y Hg A1).
Recent Medieval North African/Near Eastern admixture
A 2004 study showed that E1b1b1b is found present, albeit at low levels throughout Southern Europe (ranging from 1.5% in Northern Italians, 2.2% in Central Italians, 1.6% in southern Spaniards, 3.5% in the French, 4% in the Northern Portuguese, 12.2% in the southern Portuguese and 41.2% in the genetic isolate of the Pasiegos from Cantabria). The findings of this latter study contradict a more thorough analysis Y-chromosome analysis of the Iberian peninsula according to which haplogroup E1b1b1b surpasses frequencies of 10% in Southern Spain. The study points only to a very limited influence from northern Africa and the Middle East both in historic and prehistoric times. The absence of microsatellite variation suggests a very recent arrival from North Africa consistent with historical exchanges across the Mediterranean during the period of Islamic expansion, namely of Berber populations.. A study restricted to Portugal, concerning Y-chromosome lineages, revealed that "The mtDNA and Y data indicate that the Berber presence in that region dates prior to the Moorish expansion in 711 AD... Our data indicate that male Berbers, unlike sub-Saharan immigrants, constituted a long-lasting and continuous community in the country".
Haplotype V (p49/TaqI), a characteristic North African haplotype, may be also found in the Iberian peninsula, and a decreasing North-South cline of frequency clearly establishes a gene flow from North Africa towards Iberia which is also consistent with Moorish presence in the peninsula.. This North-South cline of frequency of halpotype V is to be observed throughout the Mediterranean region, ranging from frequencies of close to 50% in southern Portugal to around 10% in southern France. Similarly, the highest frequency in Italy is to be found in the southern island of Sicily (28%).
A wide ranging study (by Cruciani et al., published 2007) using 6,501 unrelated Y-chromosome samples from 81 populations found that: "Considering both these E-M78 sub-haplogroups (E-V12, E-V22, E-V65) and the E-M81 haplogroup, the contribution of northern African lineages to the entire male gene pool of Iberia (barring Pasiegos), continental Italy and Sicily can be estimated as 5.6%, 3.6%, and 6.6%, respectively."
A very recent study about Sicily (by Gaetano et al., 2008) found that "The Hg E3b1b-M81, widely diffused in northwestern African populations, is estimated to contribute to the Sicilian gene pool at a rate of 6%." and "confirms the genetic affinity between Sicily and North Africa".
According to one recent study (by Adams et al., December 2008) that analysed 1140 unrelated Y-chromosome samples in Iberia "mean North African admixture is 10.6%, with wide geographical variation, ranging from zero in Gascony to 21.7% in Northwest Castile".
In January 2009, a study by Capelli et al. that analysed 717 Spanish individuals, 659 Portuguese individuals and 915 Italian individuals found North African haplogroups frequencies at 7.7 % in Spain (ranging from 0% in Catalonia to 18.6% in Cantabria), 7.5% in Sicily, 7.1% in Portugal and 4.7% and in a region of Southern Italy (East Campania, Northwest Apulia, Lucera).
Genetic studies on Iberian populations also show that North African mitochondrial DNA sequences (haplogroup U6) and sub-Saharan sequences (Haplogroup L), although present at only low levels, are still at much higher levels than those generally observed elsewhere in Europe. Haplotype U6 have also been detected in Sicily at very low levels. It happens also to be a characteristic genetic marker of the Saami populations of Northern Scandinavia. It is difficult to ascertain that U6's presence is the consequence of Islam's expansion into Europe during the Middle Ages, particularly because it is more frequent in the north of the Iberian Peninsula rather than in the south. In smaller numbers it is also attested too in the British Islands, again in its northern and western borders. It may be a trace of a prehistoric neolithic/megalithic expansion along the Atlantic coasts from North Africa, perhaps in conjunction with seaborne trade. One subclade of U6 is particularly common among Canarian Spaniards as a result of native Guanche (proto-Berber) ancestry.
Sub Saharan Admixture admixture
Further information: Sub-Saharan DNA admixture in EuropeSub-Saharan African Y-chromosomes are much less common in Europe, for the reasons discussed above. The small presence of the Haplogroups E(xE3b) (i.e. clades of E other than E3b) and Haplogroup A in Europe is attributable to the slave trade, the Moor invasion of Spain or prehistoric migrations. Haplotype A has been detected in Yorkshire Portugal (3%), France (2.5% in a very small sample), Germany (2%), Sardinia (1.6%), Austria (0.78%), Italy (0.45%), Spain (0.42%), Greece (0.27%) Cyprus and Turkey.. By contrast, North Africans have about 5% paternal sub-Saharan admixture.
African Mitochondrial haplogroups are distributed along an west-to-east cline and a south-to-north cline. The highest frequencies are observed in the Iberian Peninsula.
Malyarchuk et al identified 8 African haplogroups in Russians, Czechs, Slovaks and Polish populations. These haplogroups included L1b, L2a, L3b, L3d and M1, of which some appeared to be of West African origin. Haplogroup L2a1a was identified as most likely having entered Europe about 10,000 years ago, possibly through the Iberian Peninsula.
Uralic, Central, and East Asian admixture
Central Asian Y Chromosomes are somewhat common in European populations. Tat-C (haplogroup 16) is a Y-chromosome lineage that originated in Central Asia and likely spread to Northeastern Europe with male Uralic hunter/gatherer migrations occurring over the last 4000 years. . Today it's found In Northern and Northeast Europe in low to high frequencies. It is found in Finland (55%), European Russia (14%), Ukraine (11%), Lithuania (47%), Estonia (37)%, Sweden (8%), Norway, (6%), Poland (4%), Germany (3%), Slovakia (3%), Denmark (2%), and Belarus (2%).
Inferences from ancient DNA
The genetic history of Europe has mostly been reconstructed from the modern populations of Europe, assuming genetic continuity. This is because of availability of data. However, a small number of ancient mtDNA analyses are available from both the historical and prehistorical periods. These have been summarised by Ellen Levy-Coffman in the Journal of Genetic Genealogy. There are some large differences in the frequencies of occurrence of the various haplogroups compared wth the modern population.
For example, mtDNA Haplogroup N1a, while presently rare (0.18%-0.3%), occurred in as many as 25% of Neolithic Europeans. . The cause of this reduction is unknown.
She concludes that the genetic profile of Europe has undergone significant transformation over time and that the modern population is not a living fossil of the ancient one. However, the very small sample sizes of the ancient DNA are a problem and more data is needed.
Footnotes
- Genes, peoples, and languages - Cavalli-Sforza 94 (15): 7719 - Proceedings of the National Academy of Sciences
- ^ Genes, Peoples, and Languages By L. L. (Luigi Luca) Cavalli-Sforza ISBN 0520228731
- First Chapter: 'Genes, Peoples, and Languages'
- Uralic genes in Europe by Guglielmino CR, Piazza A, Menozzi P, Cavalli-Sforza LL
- See for example Stephen Oppenheimer, Out of Eden - the peopling of the World, or Brian Sykes, The Seven Daughters of Eve.
- Seldin MF, Shigeta R, Villoslada P, Selmi C, Tuomilehto J, et al. (2006) European Population Substructure: Clustering of Northern and Southern Populations . PLoS Genet 2(9): e143. doi:10.1371/journal.pgen.0020143
- Measuring European Population Stratification using Microarray Genotype Data
- John Novembre, Toby Johnson, Katarzyna Bryc, Zoltán Kutalik, Adam R. Boyko, Adam Auton, Amit Indap, Karen S. King, Sven Bergmann, Matthew R. Nelson, Matthew Stephens & Carlos D. Bustamante (2008) "Genes mirror geography within Europe". Nature '456,' 98-101 doi:10.1038/nature07331
- "Correlation between Genetic and Geographic Structure in Europe" Oscar Lao, Timothy T. Lu, Michael Nothnagel, Olaf Junge, Sandra Freitag-Wolf, Amke Caliebe, Miroslava Balascakova, Jaume Bertranpetit, Laurence A. Bindoff, David Comas, Gunilla Holmlund, Anastasia Kouvatsi, Milan Macek, Isabelle Mollet, Walther Parson, Jukka Palo, Rafal Ploski, Antti Sajantila, Adriano Tagliabracci, Ulrik Gether, Thomas Werge, Fernando Rivadeneira, Albert Hofman, André G. Uitterlinden, Christian Gieger, Heinz-Erich Wichmann, Andreas Rüther, Stefan Schreiber, Christian Becker, Peter Nürnberg, Matthew R. Nelson, Michael Krawczak and Manfred Kayser (2008) Current Biology 18, 1241–1248, doi:10.1016/j.cub.2008.07.049
- ^ McDonald, World Haplogroups Maps
- Sykes 2006, p. 280 harvnb error: no target: CITEREFSykes2006 (help)
- Sykes 2006, pp. 281–282 harvnb error: no target: CITEREFSykes2006 (help)
- Sykes 2006, p. 283-284 harvnb error: no target: CITEREFSykes2006 (help)
- ^ DNA Heritage
- Semino et al (2000),The Genetic Legacy of Paleolithic Homo sapiens sapiens in Extant Europeans, Science Vol 290. Note: Haplogroup names are different in this article. For example: Haplogroup I is referred as M170
- World haplogroup maps
- Y-chromosome DNA Haplogroups
- Oxford Journals
- Variations of R1b Ydna in Europe: Distribution and Origins
- Y-DNA Haplogroup Tree 2006
- Y-DNA Haplogroup R and its Subclades
- Y-DNA Haplogroup I and its Subclades
- "Population genetics: DNAs from the European Neolithic".
- Cruciani et al. (2007) Tracing Past Human Male Movements in Northern/Eastern Africa and Western Eurasia: New Clues from Y-Chromosomal Haplogroups E-M78 and J-M12, Molecular Biology and Evolution, 24:1300-1311.
- Oleg Balanovsky. Two Sources of the Russian Patrilineal Heritage in Their Eurasian Context
- World mtDNA haplogroup map
- Mitochondrial DNA sequence variation in human populations, Oulu University Library (Finland)
- Coastline sketched from Mithen (2003) pp. 108-109, Extent of refugia infered from Oppenheimer (2006) p. 103.
- Richard G. Klein (March 2003). "Paleoanthropology: Whither the Neanderthals?". Science 299 (5612): 1525-1527. DOI:10.1126/science.1082025.
- Tracing European founder lineages in the Near Eastern mitochonrial gene pool, American Journal of Human Genetics, 67 pp 1252-76
- Antonio Torroni et al. "A Signal, from human mtDNA, of Postgalacian Recolonization in Europe, Am. J. Human Gen.69:844-852 (2001)
- Ornella Semin et al. The Genetic Legacy of Paleolithic Homo Sapiens Sapiens in Extant Europeans: A Y Chromosome Perspective Science 290:1155-1159, 2000.
- Tracing European founder lineages in the Near Eastern mitochondrial gene pool, Am. J. of Human Genetics, 67, 1251
- The Origins of the British
- such as Bellwood and Renfrew
- Nicholas Wade, "Before the Dawn", ch. 10. ISBN 1594200793
- Cavalli-Sforza, Luigi Luca, Paolo Menozzi, and Alberto Piazza. (1994). The History and Geography of Human Genes. Princeton University Press. ISBN 0-691-08750-4.
- Oppenheimer
- Richards
- Semino et al. (2004), Origin, Diffusion, and Differentiation of Y-Chromosome Haplogroups E and J: Inferences on the Neolithization of Europe and Later Migratory Events in the Mediterranean Area, American Journal of Human Genetics, 74: 1023–1034.
- Y-DNA Haplogroup E and its Subclades - 2008
- ^ * Semino et al. (2004) Origin, Diffusion, and Differentiation of Y-Chromosome Haplogroups E and J: Inferences on the Neolithization of Europe and Later Migratory Events in the Mediterranean Area
- See Bryan Sykes, The Seven Daughters of Eve, 1st American ed. (New York: Norton, 2001) for an entertaining account of how this consensus was reached. For historical reasons, in the 1980s mtDNA researchers believed that the Indo-European expansion was overwhelmingly a spread of technology and language, not of genes, while those who studied Y-chromosome lineages believed the opposite. Gradually the mtDNA researchers (Sykes) admitted more physical migration into their scenarios, while the Y folks (Peter Underhill) accepted more technology-copying. Eventually, both groups independently reached a 20% Neolithic - 80% Paleolithic ratio of genetic contribution to today's European population. The mtDNA vs. Y-chromosome discrepancy may be explained by noting that in such conquest-based migrations, a common pattern is of invading foreign males producing offspring with indigenous females, though significant numbers of females of the spreading culture could also arrive with post-conquest settlers. However, where migrations are essentially economic (as most migrations appear to be) it appears equally probable that male family members preceded females into new territory looking for opportunities.
- ^ King; et al. (2007). "Africans in Yorkshire?".
{{cite journal}}: Cite journal requires|journal=(help); Explicit use of et al. in:|last=(help) - Cruciani et al, 2004, Phylogeography of the Y-Chromosome Haplogroup E3b
- Reduced Genetic Structure for Iberian Peninsula: implications for population demography. (2004)
- Y-chromosome Lineages from Portugal, Madeira and Açores Record Elements of Sephardim and Berber Ancestry
- Tracing Past Human Male Movements in Northern/Eastern Africa and Western Eurasia: New Clues from Y-chromosomal Haplogroups E-M78 and J-M12
- Fluvio Cruciani, Et al. ,"Tracing Past Human Male Movements in Northern/Eastern Africa and Eurasia: New Clues from Y-Chromosomal Haplogroups E-M78 and J-M12", Molecular Biology and Evolution, Volume 24, Number 6: June 2007, Oxford University Press, Pp. 1307
- "The co-occurrence of the Berber E3b1b-M81 (2.12%) and of the Mid-Eastern J1-M267 (3.81%) Hgs together with the presence of E3b1a1-V12, E3b1a3-V22, E3b1a4-V65 (5.5%) support the hypothesis of intrusion of North African genes. (...) These Hgs are common in northern Africa and are observed only in Mediterranean Europe and together the presence of the E3b1b-M81 highlights the genetic relationships between northern Africa and Sicily. (...) Hg E3b1b-M81 network cluster confirms the genetic affinity between Sicily and North Africa.", Differential Greek and northern African migrations to Sicily are supported by genetic evidence from the Y chromosome, Gaetano et al. 2008
- The Genetic Legacy of Religious Diversity and Intolerance: Paternal Lineages of Christians, Jews, and Muslims in the Iberian Peninsula, Adams et al. 2008
- Gene Test Shows Spain’s Jewish and Muslim Mix, The New York Times, December 4, 2008
- Moors and Saracens in Europe estimating the medieval North African male legacy in southern Europe, Capelli et al, European Journal of Human Genetics, 21 January 2009
- "Haplogroup U6 is present at frequencies ranging from 0 to 7% in the various Iberian populations, with an average of 1.8%. Given that the frequency of U6 in NW Africa is 10%, the mtDNA contribution of NW Africa to Iberia can be estimated at 18%. This is larger than the contribution estimated with Y-chromosomal lineages (7%) (Bosch et al. 2001)."Joining the Pillars of Hercules: mtDNA Sequences Show Multidirectional Gene Flow in the Western Mediterranean (2003)
- "Although the absolute value of observed U6 frequency in Iberia is low, it reveals a considerable North African female contribution, if we keep in mind that haplogroup U6 is not very common in North Africa itself and virtually absent in the rest of Europe. Indeed, because the range of variation in western North Africa is 4-28%, the estimated minimum input is 8.54%"African female heritage in Iberia: a reassessment of mtDNA lineage distribution in present times (2005)
- "Our results clearly reinforce, extend, and clarify the preliminary clues of an "important mtDNA contribution from northwest Africa into the Iberian Peninsula" (Côrte-Real et al., 1996; Rando et al., 1998; Flores et al., 2000a; Rocha et al., 1999)(...) Our own data allow us to make minimal estimates of the maternal African pre-Neolithic, Neolithic, and/or recent slave trade input into Iberia. For the former, we consider only the mean value of the U6 frequency in northern African populations, excluding Saharans, Tuareg, and Mauritanians (16%), as the pre-Neolithic frequency in that area, and the present frequency in the whole Iberian Peninsula (2.3%) as the result of the northwest African gene flow at that time. The value obtained (14%) could be as high as 35% using the data of Corte-Real et al. (1996), or 27% with our north Portugal sample." Mitochondrial DNA affinities at the Atlantic fringe of Europe (2003)
- Moors and Saracens in Europe: estimating the medieval North African male legacy in southern Europe
- Sanchez et al. (2005). "High frequencies of Y chromosome lineages characterized by E3b1, DYS19-11, DYS392-12 in Somali males". European Journal of Human Genetics; 13:856–866
- Cruciani et al. 2004, Flores et al. 2004, Brion et al. 2005, Brion et al. 2004, Rosser et al. 2000, Semino et al. 2004, and DiGiacomo et al. 2003 Bosch et al. 2001 High-Resolution Analysis of Human Y-Chromosome Variation Shows a Sharp Discontinuity and Limited Gene Flow between Northwestern Africa and the Iberian Peninsula
- Malyarchuk; et al. (2008). "Reconstructing the phylogeny of African mitochondrial DNA lineages in Slavs". doi:10.1038/ejhg.2008.70.
{{cite journal}}: Cite journal requires|journal=(help); Explicit use of et al. in:|last=(help) - The Dual Origin and Siberian Affinities of Native American Y Chromosomes The American Journal of Human Genetics, 2002 January; 70(1): 192–206.
- "The network of Tat-C and DYS7C haplotypes revealed that the ancestral Tat-C haplotype (7C) was found only in southern Middle Siberia, indicating that this Y-chromosome lineage arose in that region. Moreover, the limited microsatellite diversity and resulting compact nature of the network indicates that the Tat-C lineage arose relatively recently (Zerjal et al. 1997). The absence of the Tat-C haplogroup in the Americas, with the exception of a single Navajo (Karafet et al. 1999), along with its high frequency in both northern Europe and northeastern Siberia, indicates that the Tat-C lineage was disseminated from central Asia by both westward and eastward male migrations, the eastward migration reaching Chukotka after the Bering Land Bridge was submerged. Both the M45 and Tat-C haplogroups have been found in Europe, indicating both ancient and recent central Asian influences. However, neither of these major Middle Siberian Y-chromosome lineages appears to have been greatly influenced by the paternal gene pool of Han Chinese or other East Asian populations (Su et al. 1999)."The Dual Origin and Siberian Affinities of Native American Y Chromosomes
- Dual Origins of Finns Revealed by Y Chromosome Haplotype Variation The American Journal of Human Genetics, Volume 62, Issue 5, 1171-1179, 1 May 1998
- Different genetic components in the Norwegian population revealed by the analysis of mtDNA and Y chromosome polymorphisms European Journal Of Human Genetics. September 2002, Volume 10, Number 9, Pages 521-529
- Haak, Wolfgang, et al. "Ancient DNA from the First European Farmers in 7500-Year-Old Neolithic Sites" Science, vol. 310, pg. 1016 (2005)
- Balter, Michael "Ancient DNA Yields Clues to the Puzzle of European Origins" Science, vol. 310, pg. 964 (2005)
See also
- Caucasoid
- European ethnic groups
- Human genetic variation
- Population genetics
- Archaeogenetics of the Near East
- White people
- Genetic history of the British Isles
External links
- International Society of Genetic Genealogy - 2009 tree of haplogroup R
- Journal of Genetic Genealogy - We Are Not Our Ancestors: Evidence for Discontinuity between Prehistoric and Modern Europeans
- Atlas of the Human Journey
- World Haplogroups Maps
- Origins of Europeans
- Genetic Structure of Human Populations.
- Haplotype R1b
- Haplogroup R1b