| Revision as of 10:17, 23 October 2013 editBQUB13-Gbosch (talk | contribs)8 editsNo edit summary← Previous edit | Latest revision as of 00:50, 31 December 2024 edit undoLizardJr8 (talk | contribs)Extended confirmed users, Pending changes reviewers, Rollbackers97,752 editsm Reverted edits by 184.170.185.164 (talk) (HG) (3.4.12)Tags: Huggle Rollback | ||
| (273 intermediate revisions by 92 users not shown) | |||
| Line 1: | Line 1: | ||
| {{Short description|Family of viruses}} | |||
| {{Taxobox | |||
| {{Use dmy dates|date=April 2017}} | |||
| | image = Polyomavirus.jpg | |||
| {{Virusbox | |||
| | image_size = 240px | |||
| | taxon = Polyomaviridae | |||
| | image_caption = ] showing a '''polyomavirus''' infected cell—large (blue) cell below-center-left. Urine cytology specimen. | |||
| | image = Polyomavirus.jpg | |||
| | virus_group = i | |||
| | image_caption = ] showing a '''polyomavirus''' infected cell—large (blue) cell below-center-left. Urine cytology specimen. | |||
| | familia = '''''Polyomaviridae''''' | |||
| | subdivision_ranks = Genera | |||
| | genus = '''''Avipolyomavirus'''''<br/> | |||
| | subdivision = | |||
| '''''Orthopolyomavirus'''''<br/> | |||
| * '']'' | |||
| '''''Wukipolyomavirus''''' | |||
| * '']'' | |||
| | subdivision_ranks = Species | |||
| * '']'' | |||
| | subdivision = '']''<br> | |||
| '']'' |
* '']'' | ||
| * '']'' | |||
| '']''<br> | |||
| * '']'' | |||
| ''] (SA12)''<br> | |||
| '']''<br> | |||
| '']''<br> | |||
| '']''<br> | |||
| '']''<br> | |||
| '']''<br> | |||
| '']''<br/> | |||
| '']''<br/> | |||
| '']''<br/> | |||
| '']''<br/> | |||
| '']''<br/> | |||
| '']''<br/> | |||
| '']''<br/> | |||
| '']''<br/> | |||
| '']''<br> | |||
| '']''<br/> | |||
| '']''<br/> | |||
| '']''<br/> | |||
| '']''<br/> | |||
| '']''<br> | |||
| '']''<br/> | |||
| '']''<br/> | |||
| '']''<br> | |||
| '']''<br> | |||
| '']''<br> | |||
| '']''<br> | |||
| '']''<br> | |||
| '']''<br> | |||
| '']''<br> | |||
| '']''<br> | |||
| '']''<br> | |||
| '']''<br> | |||
| '']''<br/> | |||
| '']''<br/> | |||
| '']'' | |||
| }} | }} | ||
| '''''Polyomaviruses''''' are ] (double-stranded DNA, ~5000 base pairs, circular genome), small (40–50 ] in diameter), and ] in shape, and do not have a ] envelope. Moreover, the genome possess early and late genes, contributing to its complex transcription program. They are potentially ] (]-causing); they often persist as latent infections in a host without causing disease, but may produce tumors in a host of a different species, or a host with an ineffective ]. The name ''polyoma'' refers to the viruses' ability to produce multiple (poly-) tumors (-oma). | |||
| '''''Polyomaviridae''''' is a family of ] whose natural ] are primarily mammals and birds.<ref>{{cite journal | vauthors = Moens U, Calvignac-Spencer S, Lauber C, Ramqvist T, Feltkamp MC, Daugherty MD, Verschoor EJ, Ehlers B | title = ICTV Virus Taxonomy Profile: ''Polyomaviridae'' | journal = The Journal of General Virology | volume = 98 | issue = 6 | pages = 1159–1160 | date = June 2017 | pmid = 28640744 | doi = 10.1099/jgv.0.000839 | pmc = 5656788 }}</ref><ref name=ICTVReport>{{cite web|title=ICTV Report ''Polyomaviridae''|url=http://www.ictv.global/report/polyomaviridae}}</ref> As of 2024, there are eight recognized genera.<ref>{{Cite web |last=taxonomy |title=Taxonomy browser (''Polyomaviridae'') |url=https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Undef&id=151341&lvl=3&lin=f&keep=1&srchmode=1&unlock |access-date=2024-03-19 |website=www.ncbi.nlm.nih.gov}}</ref> Fourteen species are known to infect humans, while others, such as ], have been identified in humans to a lesser extent.<ref name="Polyomaviridae2016">{{cite journal | vauthors = Calvignac-Spencer S, Feltkamp MC, Daugherty MD, Moens U, Ramqvist T, Johne R, Ehlers B | title = A taxonomy update for the family ''Polyomaviridae'' | journal = Archives of Virology | volume = 161 | issue = 6 | pages = 1739–50 | date = June 2016 | pmid = 26923930 | doi = 10.1007/s00705-016-2794-y | doi-access = free | hdl = 10037/13151 | hdl-access = free }}</ref><ref name="decaprio"/> Most of these viruses are very common and typically asymptomatic in most human populations studied.<ref name="gossai">{{cite journal | vauthors = Gossai A, Waterboer T, Nelson HH, Michel A, Willhauck-Fleckenstein M, Farzan SF, Hoen AG, Christensen BC, Kelsey KT, Marsit CJ, Pawlita M, Karagas MR | title = Seroepidemiology of Human Polyomaviruses in a US Population | journal = American Journal of Epidemiology | volume = 183 | issue = 1 | pages = 61–9 | date = January 2016 | pmid = 26667254 | doi = 10.1093/aje/kwv155 | pmc = 5006224 }}</ref><ref name="kean">{{cite journal | vauthors = Kean JM, Rao S, Wang M, Garcea RL | title = Seroepidemiology of human polyomaviruses | journal = PLOS Pathogens | volume = 5 | issue = 3 | pages = e1000363 | date = March 2009 | pmid = 19325891 | pmc = 2655709 | doi = 10.1371/journal.ppat.1000363 | doi-access = free }}</ref> BK virus is associated with ] in ] and non-renal solid organ transplant patients,<ref name="jamboti">{{cite journal | vauthors = Jamboti JS | title = BK virus nephropathy in renal transplant recipients | journal = Nephrology | volume = 21 | issue = 8 | pages = 647–54 | date = August 2016 | pmid = 26780694 | doi = 10.1111/nep.12728 | doi-access = free }}</ref><ref name="kuppachi">{{cite journal | vauthors = Kuppachi S, Kaur D, Holanda DG, Thomas CP | title = BK polyoma virus infection and renal disease in non-renal solid organ transplantation | journal = Clinical Kidney Journal | volume = 9 | issue = 2 | pages = 310–8 | date = April 2016 | pmid = 26985385 | pmc = 4792618 | doi = 10.1093/ckj/sfv143 }}</ref> JC virus with ],<ref name="adang">{{cite journal | vauthors = Adang L, Berger J | title = Progressive Multifocal Leukoencephalopathy | journal = F1000Research | volume = 4 | pages = 1424 | date = 2015 | pmid = 26918152 | pmc = 4754031 | doi = 10.12688/f1000research.7071.1 | doi-access = free }}</ref> and ] with ].<ref name=Feng_2008>{{cite journal | vauthors = Feng H, Shuda M, Chang Y, Moore PS | title = Clonal integration of a polyomavirus in human Merkel cell carcinoma | journal = Science | volume = 319 | issue = 5866 | pages = 1096–100 | date = February 2008 | pmid = 18202256 | pmc = 2740911 | doi = 10.1126/science.1152586 | bibcode = 2008Sci...319.1096F }}</ref> | |||
| The family ''Polyomaviridae'' used to be one of two genera within the now obsolete family '']'' (the other family being '']).'' The name ''Papovaviridae'' derived from three abbreviations: Pa for ''Papillomavirus'', Po for ''Polyomavirus'', and Va for "vacuolating". Clinically, Polyomaviridæ are relevant as they contribute to pathologies such as ] (]), ] (]), and ] (]). | |||
| ==Structure and genome== | |||
| Until recently, the ] of Polyomaviridae contained only one ] (Polyomavirus). The recent expansion of known Polyomaviruses called for reclassification of the family into 3 genera: Orthopolyomavirus, Wukipolyomavirus, and Avipolyomavirus.<ref name="Johne2011" /> | |||
| ] ], colored such that areas of the surface closer to the interior center appear blue and areas nearer to the surface appear red. Rendered from {{PDB|1SIE}}.]] | |||
| Polyomaviruses are ] ] viruses with circular ]s of around 5000 ]s. With such a small size, they are ranked among the smallest known double stranded DNA viruses.<ref name= Shackelton2006>{{cite journal | vauthors = Shackelton LA, Rambaut A, Pybus OG, Holmes EC |title= JC virus evolution and its association with human populations |journal= J Virol |date=October 2006 | volume = 80| issue = 20|pages=9928–9933|doi=10.1128/JVI.00441-06|pmid=17005670 |pmc= 1617318|doi-access=free}}</ref> | |||
| The genome is packaged in a ] of about 40-50 ] in diameter, which is ] in shape (T=7 symmetry).<ref name=ICTVReport /><ref name=viralzone>{{cite web | title = Viral Zone | url = http://viralzone.expasy.org/all_by_species/148.html | publisher = ExPASy | access-date=15 June 2015 }}</ref> The capsid is composed of 72 pentameric ]s of a protein called ], which is capable of self-assembly into a closed icosahedron;<ref name="salunke">{{cite journal | vauthors = Salunke DM, Caspar DL, Garcea RL | title = Self-assembly of purified polyomavirus capsid protein VP1 | journal = Cell | volume = 46 | issue = 6 | pages = 895–904 | date = September 1986 | pmid = 3019556 | doi = 10.1016/0092-8674(86)90071-1 | s2cid = 25800023 }}</ref> each pentamer of VP1 is associated with one molecule of one of the other two capsid proteins, ].<ref name="decaprio">{{cite journal | vauthors = DeCaprio JA, Garcea RL | title = A cornucopia of human polyomaviruses | journal = Nature Reviews. Microbiology | volume = 11 | issue = 4 | pages = 264–76 | date = April 2013 | pmid = 23474680 | pmc = 3928796 | doi = 10.1038/nrmicro2992 }}</ref> | |||
| ], a human polyomavirus. The early region is shown on the left and contains the TAg (tumor antigen) proteins; the late region is on the right and contains the capsid proteins.<ref name="pmid17480120">{{cite journal | vauthors = Gaynor AM, Nissen MD, Whiley DM, Mackay IM, Lambert SB, Wu G, Brennan DC, Storch GA, Sloots TP, Wang D | title = Identification of a novel polyomavirus from patients with acute respiratory tract infections | journal = PLOS Pathogens | volume = 3 | issue = 5 | pages = e64 | date = May 2007 | pmid = 17480120 | pmc = 1864993 | doi = 10.1371/journal.ppat.0030064 | doi-access = free }}</ref>]] | |||
| The genome of a typical polyomavirus codes for between five and nine ]s, divided into two ] regions called the early and late regions due to the time during infection in which they are transcribed. Each region is transcribed by the host cell's ] as a single ] containing multiple genes. The early region usually codes for two proteins, the small and large tumor antigens, produced by ]. The late region contains the three capsid structural proteins VP1, VP2, and VP3, produced by alternative ] start sites. Additional genes and other variations on this theme are present in some viruses: for example, rodent polyomaviruses have a third protein called ] in the early region, which is extremely efficient at inducing ]; ] has an additional capsid protein VP4; some examples have an additional regulatory protein called ] expressed from the late region. The genome also contains a ] control or regulatory region containing the early and late regions' ], transcriptional start sites, and the ].<ref name=ICTVReport /><ref name=viralzone /><ref name=decaprio /><ref name=iarc>{{Cite journal|last=International Agency for Research on Cancer|year=2013|title=Introduction to Polyomaviruses|url=https://monographs.iarc.fr/ENG/Monographs/vol104/mono104-0GI.pdf|journal=IARC Monographs on the Evaluation of Carcinogenic Risks to Humans|volume=104|pages=121–131}}</ref> | |||
| == Replication and life cycle == | |||
| ] was the first polyomavirus discovered by ] in 1953.<ref>{{cite journal |author=GROSS L |title=A filterable agent, recovered from Ak leukemic extracts, causing salivary gland carcinomas in C3H mice |journal=Proceedings of the Society for Experimental Biology and Medicine. Society for Experimental Biology and Medicine |volume=83 |issue=2 |pages=414–21 |year=1953 |month=June |pmid=13064287}}</ref> Subsequently, many polyomaviruses have been found to infect birds and mammals. | |||
| ] VP1 in complex with the GT1a ]. GT1a is shown in yellow and the VP1 monomer with a white surface and a blue protein backbone. A complex network of ]s, many water-mediated, is shown at the binding surface by orange lines, with participating protein residues shown as sticks. Mutations of the two residues shown in cyan at the bottom of the figure can significantly affect pathogenicity. From {{PDB|5CPW}}.<ref name=buch />]] | |||
| The polyomavirus life cycle begins with entry into a ]. Cellular receptors for polyomaviruses are ] residues of ]s, commonly ]s. The attachment of polyomaviruses to host cells is mediated by the binding of ] to sialylated glycans on the cell surface.<ref name=ICTVReport /><ref name=viralzone /><ref name=iarc /><ref name="buch">{{cite journal | vauthors = Buch MH, Liaci AM, O'Hara SD, Garcea RL, Neu U, Stehle T | title = Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity | journal = PLOS Pathogens | volume = 11 | issue = 10 | pages = e1005104 | date = October 2015 | pmid = 26474293 | pmc = 4608799 | doi = 10.1371/journal.ppat.1005104 | doi-access = free }}</ref> In some particular viruses, additional cell-surface interactions occur; for example, the ] is believed to require interaction with the ] and the ] with ].<ref name=iarc /><ref>{{cite journal | vauthors = Schowalter RM, Pastrana DV, Buck CB | title = Glycosaminoglycans and sialylated glycans sequentially facilitate Merkel cell polyomavirus infectious entry | journal = PLOS Pathogens | volume = 7 | issue = 7 | pages = e1002161 | date = July 2011 | pmid = 21829355 | pmc = 3145800 | doi = 10.1371/journal.ppat.1002161 | doi-access = free }}</ref> However, in general virus-cell interactions are mediated by commonly occurring molecules on the cell surface, and therefore are likely not a major contributor to individual viruses' observed cell-type ].<ref name=iarc /> After binding to molecules on the cell surface, the virion is ] and enters the ] - a behavior unique among known non-enveloped viruses<ref name="inoue_2013">{{cite journal | vauthors = Inoue T, Tsai B | title = How viruses use the endoplasmic reticulum for entry, replication, and assembly | journal = Cold Spring Harbor Perspectives in Biology | volume = 5 | issue = 1 | pages = a013250 | date = January 2013 | pmid = 23284050 | pmc = 3579393 | doi = 10.1101/cshperspect.a013250 }}</ref> - where the viral capsid structure is likely to be disrupted by action of host cell ] enzymes.<ref name=ICTVReport /><ref name=viralzone /><ref name="gjoerup_2010">{{cite journal | vauthors = Gjoerup O, Chang Y | title = Update on human polyomaviruses and cancer | journal = Advances in Cancer Research | volume = 106 | pages = 1–51 | date = 2010 | pmid = 20399955 | doi = 10.1016/S0065-230X(10)06001-X | isbn = 9780123747716 }}</ref> | |||
| For nearly 40 years, only two polyomaviruses were known to infect humans. Genome sequencing technologies have recently discovered seven additional human polyomaviruses, including one causing most cases of ] and another associated with transplant-associated dysplasia (TSV), that are natural infections of humans. Discovery of these polyomaviruses and other new—but previously undiscovered—viruses may provide clues to the etiologies for human diseases. | |||
| The details of transit to the nucleus are not clear and may vary among individual polyomaviruses. It has been frequently reported that an intact, albeit distorted, virion particle is released from the endoplasmic reticulum into the cell cytoplasm, where the genome is released from the capsid, possibly due to the low ] concentration in the cytoplasm.<ref name=inoue_2013 /> Both expression of viral genes and replication of the viral genome occur in the ] using host cell machinery. The early genes - comprising at minimum the ] (ST) and ] (LT) - are expressed first, from a single ] ] strand. These proteins serve to manipulate the host's ] - dysregulating the transition from ] to ], when the host cell's genome is replicated - because host cell DNA replication machinery is needed for viral genome replication.<ref name=ICTVReport /><ref name=viralzone /><ref name=iarc /> The precise mechanism of this dysregulation depends on the virus; for example, ] LT can directly bind host cell ], but ] LT does not.<ref name="andrabi_2011">{{cite journal | doi = 10.1128/JVI.05034-11 | pmid = 21835797 | pmc = 3187521 | title = Comparisons between Murine Polyomavirus and Simian Virus 40 Show Significant Differences in Small T Antigen Function | volume=85 | issue = 20 | year=2011 | journal=Journal of Virology | pages=10649–10658 | vauthors=Andrabi S, Hwang JH, Choe JK, Roberts TM, Schaffhausen BS}}</ref> LT induces DNA replication from the viral genome's non-coding control region (NCCR), after which expression of the early mRNA is reduced and expression of the late mRNA, which encodes the viral capsid proteins, begins.<ref name=gjoerup_2010 /> As these interactions begin, the LTs belonging to several polyomaviruses, including ], present oncogenic potential.<ref name = "Rotondo_2017">{{cite journal | vauthors = Rotondo JC, Bononi I, Puozzo A, Govoni M, Foschi V, Lanza G, Gafà R, Gaboriaud P, Touzé FA, Selvatici R, Martini F, Tognon M | title = Merkel Cell Carcinomas Arising in Autoimmune Disease Affected Patients Treated with Biologic Drugs, Including Anti-TNF | journal = Clinical Cancer Research | volume = 23| issue = 14 | pages = 3929–3934 | date = July 2017 | url=https://clincancerres.aacrjournals.org/content/23/14/3929| pmid = 28174236 | doi = 10.1158/1078-0432.CCR-16-2899 | doi-access = free | hdl = 11392/2378829 | hdl-access = free }}</ref> | |||
| Polyomaviruses have been extensively studied as ] in humans and animals, leading to fundamental insights into carcinogenesis, DNA replication and protein processing. The tumor suppressor molecule ] was discovered, for example, as a cellular protein bound by the major oncoprotein (cancer-causing protein) ] made by Simian vacuolating virus 40 (]). The avian polyomavirus sometimes referred to as the '']'' is a frequent cause of death among caged ]. | |||
| Several mechanisms have been described for regulating the transition from early to late gene expression, including the involvement of the LT protein in repressing the early promoter,<ref name=gjoerup_2010 /> the expression of un-terminated late mRNAs with extensions complementary to early mRNA,<ref name=iarc /> and the expression of regulatory ].<ref name=iarc /> | |||
| Expression of the late genes results in accumulation of the viral capsid proteins in the host cell cytoplasm. Capsid components enter the nucleus in order to encapsidate new viral genomic DNA. New virions may be assembled in ].<ref name=ICTVReport /><ref name=viralzone /> The mechanism of viral release from the host cell varies among polyomaviruses; some express proteins that facilitate cell exit, such as the ] or ].<ref name=gjoerup_2010 /> In some cases high levels of encapsidated virus result in cell ], releasing the virions.<ref name=iarc /> | |||
| == |
== Viral proteins == | ||
| === Tumor antigens === | |||
| ] and ] were the first to describe the polyoma virus. The virus was later named the SE Polyoma Virus in their honor. | |||
| The ] plays a key role in regulating the viral life cycle by binding to the viral origin of DNA replication where it promotes DNA synthesis. Also as the polyomavirus relies on the host cell machinery to replicate the host cell needs to be in s-phase for this to begin. Due to this, large T-antigen also modulates cellular signaling pathways to stimulate progression of the cell cycle by binding to a number of cellular control proteins.<ref>{{cite journal | vauthors = White MK, Gordon J, Reiss K, Del Valle L, Croul S, Giordano A, Darbinyan A, Khalili K | title = Human polyomaviruses and brain tumors | journal = Brain Research. Brain Research Reviews | volume = 50 | issue = 1 | pages = 69–85 | date = December 2005 | pmid = 15982744 | doi = 10.1016/j.brainresrev.2005.04.007 | s2cid = 20990837 }}</ref> This is achieved by a two prong attack of inhibiting tumor suppressing genes p53 and members of the ] (pRB) family,<ref>{{cite journal | vauthors = Kazem S, van der Meijden E, Wang RC, Rosenberg AS, Pope E, Benoit T, Fleckman P, Feltkamp MC | title = Polyomavirus-associated Trichodysplasia spinulosa involves hyperproliferation, pRB phosphorylation and upregulation of p16 and p21 | journal = PLOS ONE | volume = 9 | issue = 10 | pages = e108947 | year = 2014 | pmid = 25291363 | pmc = 4188587 | doi = 10.1371/journal.pone.0108947 | bibcode = 2014PLoSO...9j8947K | doi-access = free }}</ref> and stimulating cell growth pathways by binding cellular DNA, ATPase-helicase, DNA polymerase α association, and binding of transcription preinitiation complex factors.<ref>{{cite journal | vauthors = Kelley WL, Georgopoulos C | title = The T/t common exon of simian virus 40, JC, and BK polyomavirus T antigens can functionally replace the J-domain of the Escherichia coli DnaJ molecular chaperone | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 94 | issue = 8 | pages = 3679–84 | date = April 1997 | pmid = 9108037 | pmc = 20500 | doi = 10.1073/pnas.94.8.3679 | bibcode = 1997PNAS...94.3679K | doi-access = free }}</ref> This abnormal stimulation of the cell cycle is a powerful force for oncogenic transformation.{{citation needed|date=November 2022}} | |||
| The ] protein is also able to activate several cellular pathways that stimulate cell proliferation. Polyomavirus small T antigens commonly target protein phosphatase 2A (]),<ref>{{cite journal | vauthors = Pallas DC, Shahrik LK, Martin BL, Jaspers S, Miller TB, Brautigan DL, Roberts TM | title = Polyoma small and middle T antigens and SV40 small t antigen form stable complexes with protein phosphatase 2A | journal = Cell | volume = 60 | issue = 1 | pages = 167–76 | date = January 1990 | pmid = 2153055 | doi = 10.1016/0092-8674(90)90726-u | s2cid = 2007706 }}</ref> a key multisubunit regulator of multiple pathways including ], the mitogen-activated protein kinase (MAPK) pathway, and the stress-activated protein kinase (SAPK) pathway.<ref>{{cite journal | vauthors = Sontag E, Fedorov S, Kamibayashi C, Robbins D, Cobb M, Mumby M | title = The interaction of SV40 small tumor antigen with protein phosphatase 2A stimulates the map kinase pathway and induces cell proliferation | journal = Cell | volume = 75 | issue = 5 | pages = 887–97 | date = December 1993 | pmid = 8252625 | doi = 10.1016/0092-8674(93)90533-V | doi-access = free }}</ref><ref>{{cite journal | vauthors = Watanabe G, Howe A, Lee RJ, Albanese C, Shu IW, Karnezis AN, Zon L, Kyriakis J, Rundell K, Pestell RG | title = Induction of cyclin D1 by simian virus 40 small tumor antigen | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 93 | issue = 23 | pages = 12861–6 | date = November 1996 | pmid = 8917510 | pmc = 24011 | doi = 10.1073/pnas.93.23.12861 | bibcode = 1996PNAS...9312861W | doi-access = free }}</ref> ] small T antigen encodes a unique domain, called the LT-stabilization domain (LSD), that binds to and inhibits the ] ] regulating both cellular and viral oncoproteins.<ref>{{cite journal | vauthors = Kwun HJ, Shuda M, Feng H, Camacho CJ, Moore PS, Chang Y | title = Merkel cell polyomavirus small T antigen controls viral replication and oncoprotein expression by targeting the cellular ubiquitin ligase SCFFbw7 | journal = Cell Host & Microbe | volume = 14 | issue = 2 | pages = 125–35 | date = August 2013 | pmid = 23954152 | pmc = 3764649 | doi = 10.1016/j.chom.2013.06.008 }}</ref> Unlike for SV40, the MCV small T antigen directly transforms rodent cells in vitro.<ref>{{cite journal | vauthors = Shuda M, Kwun HJ, Feng H, Chang Y, Moore PS | title = Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator | journal = The Journal of Clinical Investigation | volume = 121 | issue = 9 | pages = 3623–34 | date = September 2011 | pmid = 21841310 | pmc = 3163959 | doi = 10.1172/JCI46323 }}</ref> | |||
| == Classification == | |||
| The classification of Polyomaviruses is constantly evolving due to the explosion of newly discovered viruses. Previously, the family of Polyomaviridae was divided into three major clades (genetically-related groups)—the SV40 clade, the avian clade, and the murine polyomavirus clade:<ref>{{cite journal |author=Pérez-Losada M, Christensen RG, McClellan DA, ''et al.'' |title=Comparing phylogenetic codivergence between polyomaviruses and their hosts |journal=Journal of Virology |volume=80 |issue=12 |pages=5663–9 |year=2006 |month=June |pmid=16731904 |pmc=1472594 |doi=10.1128/JVI.00056-06}}</ref> Recent reclassification by the ] (ICTV) recommended dividing the family of Polyomaviridae into three genera:<ref name="Johne2011">{{cite journal|last=Johne|first=R|coauthors=Buck CB, Allander T, Atwood WJ, et al|title=Taxonomical developments in the family Polyomaviridae|journal=Arch Virol|year=2011|volume=156|issue=9|pages=1627–1634|pmid=21562881|doi=10.1007/s00705-011-1008-x}}</ref> | |||
| *Genus Orthopolyomavirus (type species SV40) | |||
| *Genus Wukipolyomavirus (type species KI polyomavirus) | |||
| *Genus Avipolyomavirus (type species Avian polyomavirus) | |||
| Many of the known viruses have not been fully classified or have not yet been officially accepted; hence, the taxonomy of this family is on going. | |||
| The ] is used in ]s developed to study cancer, such as the ] system where middle T is coupled to the ] ]. There it functions as an ], while the tissue where the tumor develops is determined by the MMTV promoter.{{citation needed|date=November 2022}} | |||
| ===Unclassified viruses=== | |||
| === Capsid proteins === | |||
| A further 12 putative species have been identified in bats.<ref name=Tao2012>Tao Y, Mang S, Conrardy C, Kuzmin IV, Recuenco S, Agwanda B, Alvarez DA, Ellison JA, Gilbert AT, Moran D, Niezgoda M, J Lindblade KA, Holmes EC, Breiman RF, Rupprecht CE, Tong S (2012) Discovery of diverse Polyomaviruses in bats and the evolutionary history of the Polyomaviridae. J Gen Virol</ref> These await classification. | |||
| The polyomavirus capsid consists of one major component, ], and one or two minor components, ]. VP1 ]s form the closed ] ], and in the interior of the capsid each pentamer is associated with one molecule of either VP2 or VP3.<ref name=decaprio /><ref name=chen_1998>{{cite journal | vauthors = Chen XS, Stehle T, Harrison SC | title = Interaction of polyomavirus internal protein VP2 with the major capsid protein VP1 and implications for participation of VP2 in viral entry | language = en | journal = The EMBO Journal | volume = 17 | issue = 12 | pages = 3233–40 | date = June 1998 | pmid = 9628860 | pmc = 1170661 | doi = 10.1093/emboj/17.12.3233 }}</ref> Some polyomaviruses, such as ], do not encode or express VP3.<ref name=schowalter /> The capsid proteins are expressed from the late region of the genome.<ref name=decaprio /> | |||
| === Agnoprotein === | |||
| A polyoma virus has been isolated from horses.<ref name=Renshaw2012>Renshaw RW, Wise AG, Maes RK, Dubovi EJ. | |||
| The ] is a small multifunctional phospho-protein found in the late coding part of the genome of some polyomaviruses, most notably ], ], and ]. It is essential for proliferation in the viruses that express it and is thought to be involved in regulating the viral life cycle, particularly replication and viral exit from the host cell, but the exact mechanisms are unclear.<ref name=Sariyer2011>{{cite journal | vauthors = Sariyer IK, Saribas AS, White MK, Safak M | title = Infection by agnoprotein-negative mutants of polyomavirus JC and SV40 results in the release of virions that are mostly deficient in DNA content | journal = Virology Journal | volume = 8 | pages = 255 | date = May 2011 | pmid = 21609431 | pmc = 3127838 | doi = 10.1186/1743-422X-8-255 | doi-access = free }}</ref><ref name="saribas_2016">{{cite journal | vauthors = Saribas AS, Coric P, Hamazaspyan A, Davis W, Axman R, White MK, Abou-Gharbia M, Childers W, Condra JH, Bouaziz S, Safak M | title = Emerging From the Unknown: Structural and Functional Features of Agnoprotein of Polyomaviruses | journal = Journal of Cellular Physiology | volume = 231 | issue = 10 | pages = 2115–27 | date = October 2016 | pmid = 26831433 | doi = 10.1002/jcp.25329 | pmc = 5217748 }}</ref> | |||
| Complete genome sequence of a polyomavirus isolated from horses. J Virol 86(16):8903</ref> This virus appears to be related to the human and other primate polyoma viruses. | |||
| ==Taxonomy== | |||
| Two further polyoma viruses have been isolated from humans—STL polyomavirus and MW polyoma virus.<ref name=Lim2012>Lim ES, Reyes A, Antonio M, Saha D, Ikumapayi UN, Adeyemi M, Stine OC, Skelton R, Brennan DC, Mkakosya RS, Manary MJ, Gordon JI, Wang D (2012) Discovery of STL polyomavirus, a polyomavirus of ancestral recombinant origin that encodes a unique T antigen by alternative splicing. Virology pii: S0042-6822(12)00597-1. doi: 10.1016/j.virol.2012.12.005</ref><ref name=Siebrasse2012>Siebrasse EA, Reyes A, Lim ES, Zhao G, Mkakosya RS, Manary MJ, Gordon JI, Wang D (2012) Identification of MW polyomavirus, a novel polyomavirus in human stool. J Virol 86(19):10321–6. doi: 10.1128/JVI.01210-12</ref> | |||
| The polyomaviruses are members of group I (dsDNA viruses). The classification of polyomaviruses has been the subject of several proposed revisions as new members of the group are discovered. Formerly, polyomaviruses and ]es, which share many structural features but have very different genomic organizations, were classified together in the now-obsolete family '']''.<ref name=ICTVTaxononmy>{{cite web|title=ICTV Taxonomy Website|url=https://ictv.global/taxonomy }}</ref> (The name ''Papovaviridae'' derived from three abbreviations: Pa for ''Papillomavirus'', Po for ''Polyomavirus'', and Va for "vacuolating.")<ref name="IARC">{{cite journal|last1=International Agency for Research on Cancer|title=IARC Working Group on the Evaluation of Carcinogenic Risk to Humans. Malaria and Some Polyomaviruses (SV40, BK, JC, and Merkel Cell Viruses).|journal=IARC Monographs on the Evaluation of Carcinogenic Risks to Humans|date=2013|volume=104|url=https://www.ncbi.nlm.nih.gov/books/NBK294248/}}</ref> The polyomaviruses were divided into three major ]s (that is, genetically related groups): the SV40 clade, the avian clade, and the murine polyomavirus clade.<ref>{{cite journal | vauthors = Pérez-Losada M, Christensen RG, McClellan DA, Adams BJ, Viscidi RP, Demma JC, Crandall KA | title = Comparing phylogenetic codivergence between polyomaviruses and their hosts | journal = Journal of Virology | volume = 80 | issue = 12 | pages = 5663–9 | date = June 2006 | pmid = 16731904 | pmc = 1472594 | doi = 10.1128/JVI.00056-06 }}</ref> A subsequent proposed reclassification by the ] (ICTV) recommended dividing the family of ''Polyomaviridae'' into three genera:<ref name="Johne2011">{{cite journal | vauthors = Johne R, Buck CB, Allander T, Atwood WJ, Garcea RL, Imperiale MJ, Major EO, Ramqvist T, Norkin LC | title = Taxonomical developments in the family ''Polyomaviridae'' | journal = Archives of Virology | volume = 156 | issue = 9 | pages = 1627–34 | date = September 2011 | pmid = 21562881 | pmc = 3815707 | doi = 10.1007/s00705-011-1008-x }}</ref> | |||
| * Genus ''Orthopolyomavirus'' (type species ]) | |||
| * Genus ''Wukipolyomavirus'' (type species ]) | |||
| * Genus ''Avipolyomavirus'' (type species ]) | |||
| The current ICTV classification system recognises six genera and 117 species, of which five could not be assigned a genus. This system retains the distinction between avian and mammalian viruses, grouping the avian subset into the genus ''Gammapolyomavirus''. The six genera are:<ref name="ictv2020">{{cite web |date=March 2021 |title=Virus Taxonomy: 2020 Release |url=https://ictv.global/taxonomy |access-date=10 May 2021 |publisher=International Committee on Taxonomy of Viruses (ICTV)}}</ref> | |||
| A new species has been identified from vervet monkeys.<ref name=Yamaguchi2013>Yamaguchi H, Kobayashi S, Ishii A, Ogawa H, Nakamura I, Moonga L, Hang'ombe BM, Mweene AS, Thomas Y, Kimura T, Sawa H, Orba Y (2013) Identification of a novel polyomavirus from vervet monkeys in Zambia. J Gen Virol</ref> | |||
| * '']'' | |||
| * '']'' | |||
| * '']'' | |||
| * '']'' | |||
| * '']'' | |||
| * '']'' | |||
| The following species are unassigned to a genus:<ref name=ictv2020 /> | |||
| ===Taxonomy=== | |||
| * '']'' | |||
| * '']'' | |||
| * '']'' | |||
| * '']'' | |||
| * '']'' | |||
| Description of additional viruses is ongoing. These include the sea otter polyomavirus 1<ref name=Siqueira2017>{{cite journal | vauthors = Siqueira JD, Ng TF, Miller M, Li L, Deng X, Dodd E, Batac F, Delwart E | title = Endemic infection of stranded southern sea otters (Enhydra lutris nereis) with novel parvovirus, polyomavirus, and adenovirus | journal = Journal of Wildlife Diseases | volume = 53 | issue = 3 | pages = 532–542 | date = July 2017 | pmid = 28192039 | doi = 10.7589/2016-04-082 | s2cid = 46740584 }}</ref> and Alpaca polyomavirus<ref name=Dela2017>{{cite journal | vauthors = Dela Cruz FN, Li L, Delwart E, Pesavento PA | title = A novel pulmonary polyomavirus in alpacas (Vicugna pacos) | journal = Veterinary Microbiology | volume = 201 | pages = 49–55 | year = 2017 | pmid = 28284622 | doi = 10.1016/j.vetmic.2017.01.005 }}</ref> Another virus is the giant panda polyomavirus 1.<ref name=Qi2017>{{cite journal | vauthors = Qi D, Shan T, Liu Z, Deng X, Zhang Z, Bi W, Owens JR, Feng F, Zheng L, Huang F, Delwart E, Hou R, Zhang W | title = A novel polyomavirus from the nasal cavity of a giant panda (Ailuropoda melanoleuca) | journal = Virology Journal | volume = 14 | issue = 1 | pages = 207 | year = 2017 | pmid = 29078783 | pmc = 5658932 | doi = 10.1186/s12985-017-0867-5 | doi-access = free }}</ref> Another virus has been described from sigmodontine rodents.<ref name=Maia2018>{{cite journal | doi = 10.1007/s00705-018-3913-8 | pmid=29931397 | volume=163 | title=A novel polyomavirus in sigmodontine rodents from São Paulo State, Brazil | year=2018 | journal=Archives of Virology | pages=2913–2915 | vauthors=Gonçalves Motta Maia F, Marciel de Souza W, Sabino-Santos G, Jorge Fumagalli M, Modha S, Ramiro Murcia P, Tadeu Moraes Figueiredo L| issue=10 | s2cid=49351836 }}</ref> Another - tree shrew polyomavirus 1 - has been described in the tree shrew.<ref name=Liu2019>{{cite journal | vauthors = Liu P, Qiu Y, Xing C, Zhou JH, Yang WH, Wang Q, Li JY, Han X, Zhang YZ, Ge XY | year = 2019 | title = Detection and genome characterization of two novel papillomaviruses and a novel polyomavirus in tree shrew (Tupaia belangeri chinensis) in China | journal = Virol J | volume = 16 | issue = 1| page = 35 | doi=10.1186/s12985-019-1141-9| pmid = 30885224 | pmc = 6423848 | doi-access = free }}</ref> | |||
| '''Genus Orthopolyomavirus (type species SV40): | |||
| ''' | |||
| {| class="wikitable" | |||
| |- | |||
| ! Species Name !! Abbreviation | |||
| |- | |||
| | B-lymphotropic polyomavirus (formerly known as African green monkey polyomavirus, AGMPyV) || LPyV | |||
| |- | |||
| | Baboon polyomavirus 1 (formerly known as Simian Agent 12) || SA12 | |||
| |- | |||
| | Bat polyomavirus (formerly known as Myotis polyomavirus, MyPyV)|| BatPyV | |||
| |- | |||
| | BK polyomavirus|| BKPyV | |||
| |- | |||
| | Bornean orang-utan polyomavirus || OraPyV1 | |||
| |- | |||
| | Bovine polyomavirus|| BPyV | |||
| |- | |||
| | California sea lion polyomavirus|| SLPyV | |||
| |- | |||
| | Dolphin polyomavirus 1 || DolPyV | |||
| |- | |||
| | Hamster polyomavirus|| HaPyV | |||
| |- | |||
| | JC polyomavirus|| JCPyV | |||
| |- | |||
| | Merkel Cell polyomavirus|| MCPyV | |||
| |- | |||
| | Murine pneumotropic virus (formerly known as Kilham strain of Polyomavirus, Kilham virus, K virus) || MPtV | |||
| |- | |||
| | Murine polyomavirus|| MPyV | |||
| |- | |||
| | Simian virus 40 (formerly known as Simian vacuolating virus 40) || SV40 | |||
| |- | |||
| | Squirrel monkey polyomavirus|| SqPyV | |||
| |- | |||
| | Sumatran orang-utan polyomavirus|| OraPyV2 | |||
| |- | |||
| | Trichodysplasia spinuolsa-associated polyomavirus|| TSPyV | |||
| |} | |||
| == Human polyomaviruses == | |||
| '''Other related virus that may be a member of the genus Orthopolyomavirus but has not been approved as a species: | |||
| Most polyomaviruses do not infect humans. Of the polyomaviruses cataloged as of 2017, a total of 14 were known with human hosts.<ref name=Polyomaviridae2016 /> However, some polyomaviruses are associated with human disease, particularly in ] individuals. MCV is highly divergent from the other human polyomaviruses and is most closely related to murine polyomavirus. ] (TSV) is distantly related to MCV. Two viruses—HPyV6 and HPyV7—are most closely related to KI and WU viruses, while HPyV9 is most closely related to the African green monkey-derived lymphotropic polyomavirus (LPV).{{citation needed|date=November 2022}} | |||
| ''' | |||
| <br /> | |||
| ChPyV: Chimpanzee polyomavirus<br /> | |||
| A fourteenth virus has been described.<ref name=Gheit2017>{{cite journal | vauthors = Gheit T, Dutta S, Oliver J, Robitaille A, Hampras S, Combes JD, McKay-Chopin S, Le Calvez-Kelm F, Fenske N, Cherpelis B, Giuliano AR, Franceschi S, McKay J, Rollison DE, Tommasino M | title = Isolation and characterization of a novel putative human polyomavirus | journal = Virology | volume = 506 | pages = 45–54 | year = 2017 | pmid = 28342387 | doi = 10.1016/j.virol.2017.03.007 | pmc = 9265179 | doi-access = free }}</ref> Lyon IARC polyomavirus is related to raccoon polyomavirus.{{citation needed|date=November 2022}} | |||
| '''Genus Wukipolyomavirus (type species KI polyomavirus) | |||
| ''' | |||
| {| class="wikitable" | |||
| |- | |||
| ! Species Name !! Abbreviation | |||
| |- | |||
| | Human polyomavirus 6|| HPyV6 | |||
| |- | |||
| | Human polyomavirus 7|| HPyV7 | |||
| |- | |||
| | KI polyomavirus (formerly known as Karolinska Institute polyomavirus)|| KIPyV | |||
| |- | |||
| | WU polyomavirus (formerly known as Washington University polyomavirus)|| WUPyV | |||
| |} | |||
| ===List of human polyomaviruses=== | |||
| '''Genus Avipolyomavirus (type species Avian polyomavirus): | |||
| The following 14 polyomaviruses with human hosts had been identified and had their ]s sequenced as of 2017:<ref name=Polyomaviridae2016 /> | |||
| ''' | |||
| <br /> | |||
| {| class="wikitable" | {| class="wikitable sortable" | ||
| |- | |- | ||
| ! data-sort-type="number" | Species !! Proposed genus !! Virus name !! Abbreviation !! ] RefSeq !! Year of discovery !! Clinical correlate (if any) !! References | |||
| ! Species Name !! Abbreviation | |||
| |- | |- | ||
| | {{sort|5|''Human polyomavirus 5''}} || Alpha || ] || MCPyV || || 2008 || ]<ref name=decaprio /> || <ref name="Merkel">{{cite news|url=https://www.nytimes.com/2008/01/18/health/research/18virus.html |title=Virus Is Linked to a Powerful Skin Cancer |work=The New York Times |access-date=2008-01-18 |date=2008-01-18 |first=Lawreence K. |last=Altman | name-list-style = vanc }}</ref><ref name="Feng_2008"/><ref>{{cite journal | vauthors = Shuda M, Arora R, Kwun HJ, Feng H, Sarid R, Fernández-Figueras MT, Tolstov Y, Gjoerup O, Mansukhani MM, Swerdlow SH, Chaudhary PM, Kirkwood JM, Nalesnik MA, Kant JA, Weiss LM, Moore PS, Chang Y | title = Human Merkel cell polyomavirus infection I. MCV T antigen expression in Merkel cell carcinoma, lymphoid tissues and lymphoid tumors | journal = International Journal of Cancer | volume = 125 | issue = 6 | pages = 1243–9 | date = September 2009 | pmid = 19499546 | doi = 10.1002/ijc.24510 | pmc = 6388400 }}</ref> | |||
| | Avian polyomavirus (formerly known as Budgerigar Fledgling disease Polyomavirus, BFPyV)|| APyV | |||
| |- | |- | ||
| | {{sort|8|''Human polyomavirus 8''}} || Alpha || ] || TSPyV || || 2010 || ]<ref name=decaprio /> || <ref name="vdm">{{cite journal | vauthors = van der Meijden E, Janssens RW, Lauber C, Bouwes Bavinck JN, Gorbalenya AE, Feltkamp MC | title = Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient | journal = PLOS Pathogens | volume = 6 | issue = 7 | pages = e1001024 | date = July 2010 | pmid = 20686659 | pmc = 2912394 | doi = 10.1371/journal.ppat.1001024 | doi-access = free }}</ref><ref name="kazem">{{cite journal | vauthors = Kazem S, van der Meijden E, Feltkamp MC | title = The trichodysplasia spinulosa-associated polyomavirus: virological background and clinical implications | journal = ] | volume = 121 | issue = 8 | pages = 770–82 | date = August 2013 | pmid = 23593936 | doi = 10.1111/apm.12092 | s2cid = 13734654 | doi-access = free }}</ref> | |||
| | Canary polyomavirus|| CaPyV | |||
| |- | |- | ||
| | {{sort|9|''Human polyomavirus 9''}} || Alpha || ] || HPyV9 || || 2011 || None known || <ref name="scuda">{{cite journal | vauthors = Scuda N, Hofmann J, Calvignac-Spencer S, Ruprecht K, Liman P, Kühn J, Hengel H, Ehlers B | title = A novel human polyomavirus closely related to the african green monkey-derived lymphotropic polyomavirus | journal = Journal of Virology | volume = 85 | issue = 9 | pages = 4586–90 | date = May 2011 | pmid = 21307194 | pmc = 3126223 | doi = 10.1128/jvi.02602-10 }}</ref> | |||
| | Crow polyomavirus|| CPyV | |||
| |- | |- | ||
| | {{sort|12|''Human polyomavirus 12''}} || Alpha || ] || HPyV12 || || 2013 || None known || <ref name=Korup2013>{{cite journal | vauthors = Korup S, Rietscher J, Calvignac-Spencer S, Trusch F, Hofmann J, Moens U, Sauer I, Voigt S, Schmuck R, Ehlers B | title = Identification of a novel human polyomavirus in organs of the gastrointestinal tract | journal = PLOS ONE | volume = 8 | issue = 3 | pages = e58021 | year = 2013 | pmid = 23516426 | pmc = 3596337 | doi = 10.1371/journal.pone.0058021 | bibcode = 2013PLoSO...858021K | doi-access = free }}</ref> | |||
| | Finch polyomavirus|| FPyV | |||
| |- | |- | ||
| | {{sort|13|''Human polyomavirus 13''}} || Alpha || ] || NJPyV || || 2014 || None known || <ref name="mishra">{{cite journal | vauthors = Mishra N, Pereira M, Rhodes RH, An P, ], Jain K, Kapoor A, Briese T, Faust PL, Lipkin WI | title = Identification of a novel polyomavirus in a pancreatic transplant recipient with retinal blindness and vasculitic myopathy | journal = The Journal of Infectious Diseases | volume = 210 | issue = 10 | pages = 1595–9 | date = November 2014 | pmid = 24795478 | pmc = 4334791 | doi = 10.1093/infdis/jiu250 }}</ref> | |||
| | Goose Hemorrhagic polyomavirus|| GHPyV | |||
| |} | |||
| '''Other viruses with similarities to members of the family Polyomaviridae that have not been assigned to a polyomavirus species: | |||
| ''' | |||
| <br /> | |||
| {| class="wikitable" | |||
| |- | |- | ||
| | {{sort|1|''Human polyomavirus 1''}} || Beta || ] || BKPyV || || 1971 || Polyomavirus-associated ]; ]<ref name=decaprio /> || <ref name="gardner">{{cite journal | vauthors = Gardner SD, Field AM, Coleman DV, Hulme B | title = New human papovavirus (B.K.) isolated from urine after renal transplantation | journal = Lancet | volume = 1 | issue = 7712 | pages = 1253–7 | date = June 1971 | pmid = 4104714 | doi = 10.1016/s0140-6736(71)91776-4 }}</ref> | |||
| ! Species Name !! Abbreviation | |||
| |- | |- | ||
| | {{sort|2|''Human polyomavirus 2''}} || Beta || ] || JCPyV || || 1971 || ]<ref name=decaprio /> || <ref name="padgett">{{cite journal | vauthors = Padgett BL, Walker DL, ZuRhein GM, Eckroade RJ, Dessel BH | title = Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy | journal = Lancet | volume = 1 | issue = 7712 | pages = 1257–60 | date = June 1971 | pmid = 4104715 | doi = 10.1016/S0140-6736(71)91777-6 }}</ref> | |||
| | Athymic rat polyomavirus|| RatPyV | |||
| |- | |- | ||
| | {{sort|3|''Human polyomavirus 3''}} || Beta || ] || KIPyV || || 2007 || None known || <ref name="Allander">{{cite journal | vauthors = Allander T, Andreasson K, Gupta S, Bjerkner A, Bogdanovic G, Persson MA, Dalianis T, Ramqvist T, Andersson B | title = Identification of a third human polyomavirus | journal = Journal of Virology | volume = 81 | issue = 8 | pages = 4130–6 | date = April 2007 | pmid = 17287263 | pmc = 1866148 | doi = 10.1128/JVI.00028-07 }}</ref> | |||
| | Baboon polyomavirus 2|| BPyV2 | |||
| |- | |- | ||
| | {{sort|4|''Human polyomavirus 4''}} || Beta || ] || WUPyV || || 2007 || None known || <ref name="pmid17480120"/> | |||
| | Cynomolgus polyomavirus|| CyPV | |||
| |- | |- | ||
| | {{sort|6|''Human polyomavirus 6''}} || Delta || ] || HPyV6 || || 2010 || HPyV6 associated pruritic and dyskeratotic dermatosis (H6PD)<ref name="nguyen_2016">{{cite journal | vauthors = Nguyen KD, Lee EE, Yue Y, Stork J, Pock L, North JP, Vandergriff T, Cockerell C, Hosler GA, Pastrana DV, Buck CB, Wang RC | title = Human polyomavirus 6 and 7 are associated with pruritic and dyskeratotic dermatoses | journal = Journal of the American Academy of Dermatology | volume = 76 | issue = 5 | pages = 932–940.e3 | date = May 2017 | pmid = 28040372 | pmc = 5392424 | doi = 10.1016/j.jaad.2016.11.035 | url = https://zenodo.org/record/995723 }}</ref> || <ref name=schowalter>{{cite journal | vauthors = Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB | title = Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin | journal = Cell Host & Microbe | volume = 7 | issue = 6 | pages = 509–15 | date = June 2010 | pmid = 20542254 | pmc = 2919322 | doi = 10.1016/j.chom.2010.05.006 }}</ref> | |||
| | Gorilla gorilla gorilla polyomavirus 1|| GggPyV1 | |||
| |- | |- | ||
| | {{sort|7|''Human polyomavirus 7''}} || Delta || ] || HPyV7 || || 2010 || HPyV7-related epithelial hyperplasia<ref name="nguyen_2016" /><ref name="ho_2015">{{cite journal | vauthors = Ho J, Jedrych JJ, Feng H, Natalie AA, Grandinetti L, Mirvish E, Crespo MM, Yadav D, Fasanella KE, Proksell S, Kuan SF, Pastrana DV, Buck CB, Shuda Y, Moore PS, Chang Y | title = Human polyomavirus 7-associated pruritic rash and viremia in transplant recipients | journal = The Journal of Infectious Diseases | volume = 211 | issue = 10 | pages = 1560–5 | date = May 2015 | pmid = 25231015 | pmc = 4425822 | doi = 10.1093/infdis/jiu524 }}</ref><ref name="toptan_2016">{{cite journal | vauthors = Toptan T, Yousem SA, Ho J, Matsushima Y, Stabile LP, Fernández-Figueras MT, Bhargava R, Ryo A, Moore PS, Chang Y | title = Survey for human polyomaviruses in cancer | journal = JCI Insight | volume = 1 | issue = 2 | date = February 2016 | pmid = 27034991 | pmc = 4811373 | doi = 10.1172/jci.insight.85562 }}</ref>|| <ref name=schowalter /> | |||
| | Human polyomavirus 9|| HPyV9 | |||
| |- | |- | ||
| | {{sort|10|''Human polyomavirus 10''}} || Delta || ] || MWPyV || || 2012 || None known || <ref name=Siebrasse2012>{{cite journal | vauthors = Siebrasse EA, Reyes A, Lim ES, Zhao G, Mkakosya RS, Manary MJ, Gordon JI, Wang D | title = Identification of MW polyomavirus, a novel polyomavirus in human stool | journal = Journal of Virology | volume = 86 | issue = 19 | pages = 10321–6 | date = October 2012 | pmid = 22740408 | pmc = 3457274 | doi = 10.1128/JVI.01210-12 }}</ref><ref name=Buck2012>{{cite journal | vauthors = Buck CB, Phan GQ, Raiji MT, Murphy PM, McDermott DH, McBride AA | title = Complete genome sequence of a tenth human polyomavirus | journal = Journal of Virology | volume = 86 | issue = 19 | pages = 10887 | date = October 2012 | pmid = 22966183 | pmc = 3457262 | doi = 10.1128/JVI.01690-12 }}</ref><ref name=Yu2012>{{cite journal|author2-link=Alexander L. Greninger | vauthors = Yu G, Greninger AL, Isa P, Phan TG, Martínez MA, de la Luz Sanchez M, Contreras JF, Santos-Preciado JI, ], Miller S, DeRisi JL, Delwart E, Arias CF, Chiu CY | title = Discovery of a novel polyomavirus in acute diarrheal samples from children | journal = PLOS ONE | volume = 7 | issue = 11 | pages = e49449 | year = 2012 | pmid = 23166671 | pmc = 3498111 | doi = 10.1371/journal.pone.0049449 | bibcode = 2012PLoSO...749449Y | doi-access = free }}</ref> | |||
| | Mastomys polyomavirus (multimammate mouse – Mastomys species)|| ? | |||
| |- | |- | ||
| | {{sort|11|''Human polyomavirus 11''}} || Delta || ] ||STLPyV|| || 2013 || None known || <ref name="Lim2013">{{cite journal | vauthors = Lim ES, Reyes A, Antonio M, Saha D, Ikumapayi UN, Adeyemi M, Stine OC, Skelton R, Brennan DC, Mkakosya RS, Manary MJ, Gordon JI, Wang D | title = Discovery of STL polyomavirus, a polyomavirus of ancestral recombinant origin that encodes a unique T antigen by alternative splicing | journal = Virology | volume = 436 | issue = 2 | pages = 295–303 | date = February 2013 | pmid = 23276405 | pmc = 3693558 | doi = 10.1016/j.virol.2012.12.005 }}</ref> | |||
| | Pan troglodytes verus polyomavirus 1a || PtvPyV1a | |||
| |- | |- | ||
| | |
|''Human polyomavirus 14'' | ||
| | Alpha | |||
| |Lyon IARC polyomavirus | |||
| |LIPyV | |||
| | | |||
| |2017 | |||
| |None known | |||
| |<ref>{{Cite journal|last1=Gheit|first1=Tarik|last2=Dutta|first2=Sankhadeep|last3=Oliver|first3=Javier|last4=Robitaille|first4=Alexis|last5=Hampras|first5=Shalaka|last6=Combes|first6=Jean-Damien|last7=McKay-Chopin|first7=Sandrine|last8=Calvez-Kelm|first8=Florence Le|last9=Fenske|first9=Neil|title=Isolation and characterization of a novel putative human polyomavirus|journal=Virology|volume=506|pages=45–54|doi=10.1016/j.virol.2017.03.007|pmid=28342387|year=2017|pmc=9265179 |doi-access=free}}</ref><ref>{{cite journal | pmid = 30328951 | doi=10.6061/clinics/2018/e558s | volume=73 | title=Human polyomaviruses and cancer: an overview | pmc=6157077 | year=2018 | journal=Clinics (Sao Paulo) | page=e558s | vauthors=Prado JC, Monezi TA, Amorim AT, Lino V, Paladino A, Boccardo E| issue=suppl 1 }}</ref> | |||
| |- | |- | ||
| | Rabbit kidney vacuolating virus|| RKV | |||
| |} | |} | ||
| ''Deltapolyomavirus'' contains only the four human viruses shown in the above table. The Alpha and Beta groups contain viruses that infect a variety of mammals. The Gamma group contains the avian viruses.<ref name=Polyomaviridae2016 /> Clinically significant disease associations are shown only where causality is expected.<ref name=decaprio /><ref name="dalianis_2013">{{cite journal | vauthors = Dalianis T, Hirsch HH | title = Human polyomaviruses in disease and cancer | journal = Virology | volume = 437 | issue = 2 | pages = 63–72 | date = March 2013 | pmid = 23357733 | doi = 10.1016/j.virol.2012.12.015 | doi-access = }}</ref> | |||
| ==Genome== | |||
| Antibodies to the monkey lymphotropic polyomavirus have been detected in humans suggesting that this virus - or a closely related virus - can infect humans.<ref name=VanGhelue2012>{{cite journal | vauthors = Van Ghelue M, Khan MT, Ehlers B, Moens U | title = Genome analysis of the new human polyomaviruses | journal = Reviews in Medical Virology | volume = 22 | issue = 6 | pages = 354–77 | date = November 2012 | pmid = 22461085 | doi = 10.1002/rmv.1711 | doi-access = free }}</ref> | |||
| The genome is circular, composed of double stranded DNA and has six genes: large T, small t, viral protein 1 (VP1), viral protein 2 (VP2), and viral protein 3 (VP3) and agnoprotein. It is about 5 kilobase pairs in length. VP1-3 form the viral capsid. | |||
| ===Clinical relevance=== | |||
| == Replication == | |||
| All the polyomaviruses are highly common childhood and young adult infections.<ref name=Egli2009>{{cite journal | vauthors = Egli A, Infanti L, Dumoulin A, Buser A, Samaridis J, Stebler C, Gosert R, Hirsch HH | title = Prevalence of polyomavirus BK and JC infection and replication in 400 healthy blood donors | journal = The Journal of Infectious Diseases | volume = 199 | issue = 6 | pages = 837–46 | date = March 2009 | pmid = 19434930 | doi = 10.1086/597126 | doi-access = free }}</ref> Most of these infections appear to cause little or no symptoms. These viruses are probably lifelong persistent among almost all adults. Diseases caused by human polyomavirus infections are most common among ] people; disease associations include ] with ] in ] and non-renal solid organ transplant patients,<ref name="jamboti"/><ref name="kuppachi"/> ] with ],<ref name="adang"/> and ] (MCV) with ].<ref name="Feng_2008"/> | |||
| Prior to genome replication, the processes of viral attachment, entry and uncoating occur. Cellular receptors for polyomaviruses are sialic acid residues of gangliosides. The attachment of polyomaviruses to host cells is mediated by viral protein 1 (]) via the sialic acid attachment region. This can be confirmed as anti-VP1 antibodies have been shown to prevent the binding of polyomavirus to host cells.<ref>http://www.microbiologybytes.com/virology/Polyomaviruses.html</ref> | |||
| ====SV40==== | |||
| Polyomavirus virions are subsequently ] and transported first to the endoplasmatic reticulum where a conformational change occurs revealing Vp2. {{Citation needed|date=July 2009}} Then by an unknown mechanism the virus is exported to the nucleus.{{Citation needed|date=July 2009}} | |||
| {{Main|SV40}} | |||
| SV40 replicates in the kidneys of ]s without causing disease, but can cause cancer in rodents under laboratory conditions. In the 1950s and early 1960s, well over 100 million people may have been exposed to SV40 due to previously undetected SV40 contamination of ], prompting concern about the possibility that the virus might cause disease in humans.<ref name=pmid16963733>{{cite journal | vauthors = Poulin DL, DeCaprio JA | title = Is there a role for SV40 in human cancer? | journal = Journal of Clinical Oncology | volume = 24 | issue = 26 | pages = 4356–65 | date = September 2006 | pmid = 16963733 | doi = 10.1200/JCO.2005.03.7101 }}</ref><ref name=pmid14566815>{{cite journal | vauthors = zur Hausen H | title = SV40 in human cancers--an endless tale? | journal = International Journal of Cancer | volume = 107 | issue = 5 | pages = 687 | date = December 2003 | pmid = 14566815 | doi = 10.1002/ijc.11517 | doi-access = }}</ref> Although it has been reported as present in some human cancers, including ]s, ]s, ]s, and ]s,<ref name="gazdar">{{cite journal | vauthors = Gazdar AF, Butel JS, Carbone M | title = SV40 and human tumours: myth, association or causality? | journal = Nature Reviews. Cancer | volume = 2 | issue = 12 | pages = 957–64 | date = December 2002 | pmid = 12459734 | doi = 10.1038/nrc947 | s2cid = 8878662 }}</ref> accurate detection is often confounded by high levels of cross-reactivity for SV40 with widespread human polyomaviruses.<ref name=pmid14566815/> Most virologists dismiss SV40 as a cause for human cancers.<ref name=pmid16963733/><ref name="BJCancer35">{{cite journal | vauthors = Carroll-Pankhurst C, Engels EA, Strickler HD, Goedert JJ, Wagner J, Mortimer EA | title = Thirty-five year mortality following receipt of SV40- contaminated polio vaccine during the neonatal period | journal = British Journal of Cancer | volume = 85 | issue = 9 | pages = 1295–7 | date = November 2001 | pmid = 11720463 | pmc = 2375249 | doi = 10.1054/bjoc.2001.2065 }}</ref><ref name="shah">{{cite journal | vauthors = Shah KV | title = SV40 and human cancer: a review of recent data | journal = International Journal of Cancer | volume = 120 | issue = 2 | pages = 215–23 | date = January 2007 | pmid = 17131333 | doi = 10.1002/ijc.22425 | s2cid = 20679358 | doi-access = free }}</ref> | |||
| === Diagnosis === | |||
| Polyomaviruses replicate in the ] of the host. They are able to utilise the host’s machinery because the ] structure is ] to that of the mammalian host. Moreover, the promoter sequence of Polyomavirus' ] is a strong attractant for the host's ]. Viral ] occurs in two distinct phases; early and late gene expression, separated by genome replication. | |||
| The diagnosis of polyomavirus almost always occurs after the primary infection as it is either asymptomatic or sub-clinical. Antibody assays are commonly used to detect presence of antibodies against individual viruses.<ref name="Cinthia B. Drachenberg 2005">{{cite journal | vauthors = Drachenberg CB, Hirsch HH, Ramos E, Papadimitriou JC | title = Polyomavirus disease in renal transplantation: review of pathological findings and diagnostic methods | journal = Human Pathology | volume = 36 | issue = 12 | pages = 1245–55 | date = December 2005 | pmid = 16311117 | doi = 10.1016/j.humpath.2005.08.009 }}</ref> Competition assays are frequently needed to distinguish among highly similar polyomaviruses.<ref>{{cite book |last1=Viscidi |first1=Raphael P. |last2=Clayman |first2=Barbara | name-list-style = vanc |year=2006 |chapter=Serological Cross Reactivity between Polyomavirus Capsids|chapter-url=https://books.google.com/books?id=Wz2aOQvHEPQC&pg=PA73 |pages=73–84 |doi=10.1007/0-387-32957-9_5 |pmid=16626028 |editor1-first=Nasimul |editor1-last=Ahsan |title=Polyomaviruses and Human Diseases |series=Advances in Experimental Medicine and Biology |volume=577 |isbn=978-0-387-29233-5}}</ref> | |||
| In cases of progressive multifocal leucoencephalopathy (PML), a cross-reactive antibody to SV40 T antigen (commonly Pab419) is used to stain tissues directly for the presence of JC virus T antigen. PCR can be used on a biopsy of the tissue or ] to amplify the polyomavirus DNA. This allows not only the detection of polyomavirus but also which sub type it is.<ref>{{cite journal | vauthors = Drews K, Bashir T, Dörries K | title = Quantification of human polyomavirus JC in brain tissue and cerebrospinal fluid of patients with progressive multifocal leukoencephalopathy by competitive PCR | journal = Journal of Virological Methods | volume = 84 | issue = 1 | pages = 23–36 | date = January 2000 | pmid = 10644084 | doi = 10.1016/S0166-0934(99)00128-7 }}</ref> | |||
| '''''Early ]''''' is responsible for the synthesis of non-structural ]. Since Polyomaviruses rely on the host to control both the gene expression, the role of the non-structural proteins is to regulate the cellular mechanisms. Close to the N terminal end of polyomavirus genome are enhancer elements which induce activation and transcription of a molecule known as the T-antigen (see ]). Early mRNA’s, encoding T-antigen are produced by host RNA polymerase II. T-antigen autoregulates early mRNA’s, subsequently leading to elevated levels of T-antigen. At high concentrations of T-antigen, early gene expression is repressed, triggering the late phase of viral infection to begin. | |||
| Genome replication acts to separate the early and late phase gene expression. The duplicated viral genome is synthesised and processed as if it were cellular DNA, exploiting the host’s machinery. As the daughter viral DNA are synthesised they associate with cellular ] to form structures that are often referred to as "minichromosomes". In this manner the DNA is packaged more efficiently. | |||
| '''''Late gene expression''''' synthesises the structural proteins, responsible for the viral particle composition. This occurs during and after genome replication. As with the early gene expression products, late gene expression generates an array of proteins as a result of ]. | |||
| Within each viral protein are 'nuclear localization signals' which cause the viral proteins to amass in the nucleus. Assembly of new virus particles consequently occurs within the nucleus of the host cell{{Citation needed|date=June 2010}}. | |||
| '''''Release''''' of newly synthesized polyomavirus particles exit the infected cell by one of two mechanisms. Firstly and less commonly, they are transported in cytoplasmic vacuoles to the ], where ] occurs. More frequently, they are released when the cell ] due to the ] of virus particles present in the infected cell. | |||
| == The Polyoma large and small T-Antigen == | |||
| The '''large T-antigen ''' plays a key role in regulating the viral life cycle by binding to the viral origin of DNA replication where it promotes DNA synthesis. Also as the polyomavirus relies on the host cell machinery to replicate the host cell needs to be in s-phase for this to begin. Due to this, large T-antigen also modulates cellular signaling pathways to stimulate progression of the cell cycle by binding to a number of cellular control proteins.<ref>{{cite journal |author=White MK, Gordon J, Reiss K, ''et al.'' |title=Human polyomaviruses and brain tumors |journal=Brain Research. Brain Research Reviews |volume=50 |issue=1 |pages=69–85 |year=2005 |month=December |pmid=15982744 |doi=10.1016/j.brainresrev.2005.04.007}}</ref> This is achieved by a two prong attack of inhibiting tumor suppressing genes p53 and members of the ] (pRB) family, and stimulating cell growth pathways by binding cellular DNA, ATPase-helicase, DNA polymerase α association, and binding of transcription preinitiation complex factors.<ref>{{cite journal |author=Kelley WL, Georgopoulos C |title=The T/t common exon of simian virus 40, JC, and BK polyomavirus T antigens can functionally replace the J-domain of the Escherichia coli DnaJ molecular chaperone |journal=Proceedings of the National Academy of Sciences of the United States of America |volume=94 |issue=8 |pages=3679–84 |year=1997 |month=April |pmid=9108037 |pmc=20500 |doi=10.1073/pnas.94.8.3679}}</ref> This abnormal stimulation of the cell cycle is a powerful force for oncogenic transformation. | |||
| The '''small T-antigen ''' protein is also able to activate several cellular pathways which stimulate cell proliferation. Such as the mitogen-activated protein kinase (MAPK) pathway, and the stress-activated protein kinase (SAPK) pathway.<ref>{{cite journal |author=Sontag E, Fedorov S, Kamibayashi C, Robbins D, Cobb M, Mumby M |title=The interaction of SV40 small tumor antigen with protein phosphatase 2A stimulates the map kinase pathway and induces cell proliferation |journal=Cell |volume=75 |issue=5 |pages=887–97 |year=1993 |month=December |pmid=8252625 |doi=10.1016/0092-8674(93)90533-V}}</ref><ref>{{cite journal |author=Watanabe G, Howe A, Lee RJ, ''et al.'' |title=Induction of cyclin D1 by simian virus 40 small tumor antigen |journal=Proceedings of the National Academy of Sciences of the United States of America |volume=93 |issue=23 |pages=12861–6 |year=1996 |month=November |pmid=8917510 |pmc=24011 |doi=10.1073/pnas.93.23.12861}}</ref> | |||
| == The Polyoma Middle T-Antigen == | |||
| The Polyoma Middle T-Antigen is used in animal ] like the ] where it is coupled to the ] ]. There it functions as an ], while the tissue where the tumor develops is determined by the MMTV promoter. | |||
| == Agnoprotein == | |||
| The agnoprotein is a small multifunctional phospho-protein found in the late coding part of the genome. It appears to be involved in DNA replication but the exact mechanism remains unclear.<ref name=Sariyer2011>Sariyer IK, Saribas AS, White MK, Safak M (2011) Infection by agnoprotein-negative mutants of polyomavirus JC and SV40 results in the release of virions that are mostly deficient in DNA content. Virol J. 8(1):255</ref> | |||
| == Human Polyomaviruses == | |||
| Several polyomaviruses have been found in humans. Four of these viruses (], ], ] and ]) are closely related to SV40 and infection with these viruses can be confused with SV40 infection.<ref name=pmid16963733>{{cite journal |author=Poulin DL, DeCaprio JA |title=Is there a role for SV40 in human cancer? |journal=Journal of Clinical Oncology |volume=24 |issue=26 |pages=4356–65 |year=2006 |month=September |pmid=16963733 |doi=10.1200/JCO.2005.03.7101}}</ref><ref name=pmid14566815>{{cite journal |author=zur Hausen H |title=SV40 in human cancers--an endless tale? |journal=International Journal of Cancer |volume=107 |issue=5 |pages=687 |year=2003 |month=December |pmid=14566815 |doi=10.1002/ijc.11517}}</ref> ] (MCV) is highly divergent from the other human polyomaviruses and is most closely related to murine polyomavirus. ] (TSV), is distantly related to MCV. Two viruses—HPyV6 and HPyV7—are most closely related to KI and WU viruses, while HPyV9 is most closely related to the African green monkey-derived lymphotropic polyomavirus (LPV). | |||
| ===Virus listing=== | |||
| * ] can infect the ], ], or ] (sometimes causing the fatal ] in the latter case). This virus like BK virus was described in 1971. | |||
| * ] produces a mild respiratory infection and can affect the kidneys of immunosuppressed ] patients. Both of these viruses are very widespread: approximately 80 percent of the adult population in the ] have antibodies to BK and JC. | |||
| * Two polyomaviruses, KI (Karolinska Institute)<ref name="Allander">{{cite journal |author=Allander T, Andreasson K, Gupta S, ''et al.'' |title=Identification of a third human polyomavirus |journal=Journal of Virology |volume=81 |issue=8 |pages=4130–6 |year=2007 |month=April |pmid=17287263 |pmc=1866148 |doi=10.1128/JVI.00028-07}}</ref> and WU (Washington University)<ref name="pmid17480120">{{cite journal |author=Gaynor AM, Nissen MD, Whiley DM, ''et al.'' |title=Identification of a novel polyomavirus from patients with acute respiratory tract infections |journal=PLoS Pathogens |volume=3 |issue=5 |pages=e64 |year=2007 |month=May |pmid=17480120 |pmc=1864993 |doi=10.1371/journal.ppat.0030064}}</ref> viruses, are closely related to each other and have been isolated from respiratory secretions. These viruses, discovered almost simultaneously in 2007, were the first of an expanding group of polyomaviruses found to naturally infect humans beyond JCV and BKV. | |||
| * In January 2008, a new virus, ], was described and shown to cause most ].<ref name="Merkel">{{cite news|url=http://www.nytimes.com/2008/01/18/health/research/18virus.html |title=Virus Is Linked to a Powerful Skin Cancer |publisher= New York Times |accessdate=2008-01-18 |date=2008-01-18|work=|author=L Altman}}</ref><ref>{{cite journal |author=Feng H, Shuda M, Chang Y, Moore PS |title=Clonal integration of a polyomavirus in human Merkel cell carcinoma |journal=Science |volume=319 |issue=5866 |pages=1096–100 |year=2008 |month=February |pmid=18202256 |pmc=2740911 |doi=10.1126/science.1152586}}</ref><ref>{{cite journal |author=Shuda M, Arora R, Kwun HJ, ''et al.'' |title=Human Merkel cell polyomavirus infection I. MCV T antigen expression in Merkel cell carcinoma, lymphoid tissues and lymphoid tumors |journal=International Journal of Cancer |volume=125 |issue=6 |pages=1243–9 |year=2009 |month=September |pmid=19499546 |doi=10.1002/ijc.24510}}</ref> | |||
| * In 2010, three new polyomaviruses infecting skin were discovered: HPyV6 and HPyV7:<ref>http://www.ncbi.nlm.nih.gov/pubmed/20542254?dopt=Abstract</ref> these two viruses are as yet not associated with human disease. Trichodysplasia spinulosa-associated polyomavirus (TSV) was discoved in the proliferative skin lesion termed trichodysplasia spinulosa seen in immunosuppressed patients.<ref name="plospathogens.org">http://www.plospathogens.org/article/info:doi/10.1371/journal.ppat.1001024</ref><ref name="plospathogens.org"/> All three of these viruses were discovered by ] of human skin DNA that preferentially amplifies small circular genomes, such as polyomaviruses. | |||
| * In March, 2011, a ninth polyoma virus HPyV9, related to a monkey lymphotropic virus (LPV) was cultured from the blood of immunosuppressed patients. The finding partially explains why some humans had antisera cross reactive with monkey LPV but none of the known human polyomaviruses cross-reacted with those patient's sera.<ref>http://www.ncbi.nlm.nih.gov/pubmed/7017066</ref> | |||
| * In 2012 a new polyoma virus (Malawi polyomavirus—MWPyV) was isolated from the stool of a healthy child from Malawi.<ref name=Siebrasse2012>Siebrasse EA, Reyes A, Lim ES, Zhao G, Mkakosya RS, Manary MJ, Gordon JI, Wang D (2012) Identification of MW polyomavirus, a novel polyomavirus in human stool. J Virol</ref> This virus has also been isolated in St. Louis, ]. It appears to be highly divergent from other members of this virus family. | |||
| * In 2012 another new polyoma virus—human polyoma virus 10—has been isolated from patient with the ]s, ], ]s and ] syndrome.<ref name=Buck2012>Buck CB, Phan GQ, Raiji MT, Murphy PM, McDermott DH, McBride AA (2012) Complete genome sequence of a tenth human polyomavirus. J Virol 86(19):10887</ref> | |||
| * In 2012 another polyoma virus—MX polyomavirus—was isolated from stool samples.<ref name=Yu2012>Yu G, Greninger AL, Isa P, Phan TG, Martínez MA, de la Luz Sanchez M, Contreras JF, Santos-Preciado JI, Parsonnet J, Miller S, Derisi JL, Delwart E, Arias CF, Chiu CY (2012) Discovery of a novel polyomavirus in acute diarrheal samples from children. | |||
| PLoS One 7(11):e49449. doi: 10.1371/journal.pone.0049449</ref> This virus was isolated from samples from ], ] and ]. This virus was also isolated from a respiratory tract infection in Mexico. It's potential for pathogenicity (if any) is currently unknown. | |||
| * In 2013 a new polyoma virus (Human Polyomavirus 12) was found in resected liver tissue.<ref name=Korup2013>Korup S, Rietscher J, Calvignac-Spencer S, Trusch F, Hofmann J, Moens U, Sauer I, Voigt S, Schmuck R, Ehlers B (2013) Identification of a novel human polyomavirus in organs of the gastrointestinal tract.PLoS One. 2013;8(3):e58021. doi: 10.1371/journal.pone.0058021.</ref> Subclinical infection with this virus appears to be common (10–20% of asymptomatic population). | |||
| *In 2013 a new polyoma virus—STL polyomavirus—was isolated from human faeces.<ref name=Lim2013>Lim ES, Reyes A, Antonio M, Saha D, Ikumapayi UN, Adeyemi M, Stine OC, Skelton R, Brennan DC, Mkakosya RS, Manary MJ, Gordon JI, Wang D (2013) Discovery of STL polyomavirus, a polyomavirus of ancestral recombinant origin that encodes a unique T antigen by alternative splicing. Virology 436(2):295–303. doi: 10.1016/j.virol.2012.12.005 </ref> | |||
| ==Clinical releveance== | |||
| All the polyomaviruses are highly common childhood and young adult infections.<ref name=Egli2009>Egli A, Infanti L, Dumoulin A, Buser A, Samaridis J, Stebler C, Gosert R, Hirsch HH. J Infect Dis. 2009 Mar 15;199(6):837–46.</ref> Most of these infections appear to cause little or no symptoms. These viruses are probably lifelong persistent among almost all adults. Diseases caused by human polyomavirus infections are most common among persons who become immunosuppressed by ], old age or after transplantation and include ], ] and ]. | |||
| In addition to its role in Merkel cell carcinoma, Merkel cell polyomavirus has been reported from a number of other conditions including non small cell ].,<ref name=Hashida2013>Hashida Y, Imajoh M, Nemoto Y, Kamioka M, Taniguchi A, Taguchi T, Kume M, Orihashi K, Daibata M (2013) Detection of Merkel cell polyomavirus with a tumour-specific signature in non-small cell lung cancer. Br J Cancer doi: 10.1038/bjc.2012.567</ref> ], ] of the skin, ] and ].<ref name=Imajoh2012>Imajoh M, Hashida Y, Nemoto Y, Oguri H, Maeda N, Furihata M, Fukaya T, Daibata M (2012) Detection of Merkel cell polyomavirus in cervical squamous cell carcinomas and adenocarcinomas from Japanese patients. Virol J 9:154. doi: 10.1186/1743-422X-9-154</ref><ref name=Murakami2011>Murakami M, Imajoh M, Ikawa T, Nakajima H, Kamioka M, Nemoto Y, Ujihara T, Uchiyama J, Matsuzaki S, Sano S, Daibata M (2011) Presence of Merkel cell polyomavirus in Japanese cutaneous squamous cell carcinoma. J Clin Virol 50(1):37–41. doi: 10.1016/j.jcv.2010.09.013</ref><ref name=Zur2009>Zur Hausen A (2009) Merkel cell polyomavirus in the pathogenesis of non-melanoma skin cancer. Pathologe 30 Suppl 2:217–20. doi: 10.1007/s00292-009-1222-4</ref><ref name=Kassem2009>Kassem A, Technau K, Kurz AK, Pantulu D, Löning M, Kayser G, Stickeler E, Weyers W, Diaz C, Werner M, Nashan D, Zur Hausen A (2009) Merkel cell polyomavirus sequences are frequently detected in nonmelanoma skin cancer of immunosuppressed patients. Int J Cancer 125(2):356–61. doi: 10.1002/ijc.24323.</ref> These putative associations awaits confirmation. It has also been associated with atypical ].<ref name=Andres2010>Andres C, Puchta U, Flaig MJ (2010) Detection of Merkel cell polyomavirus DNA in atypical fibroxanthoma in correlation to clinical features. Am J Dermatopathol 32(8):799–803. doi: 10.1097/DAD.0b013e3181dfcdff</ref> | |||
| ===SV40=== | |||
| The SV40 replicates in the kidneys of ]s without causing disease, but causes ]s in ]s. It is highly controversial whether it can cause disease in humans since the virus may have been introduced into the general population in the 1950s through a contaminated ]. Thus far, no widely-accepted evidence for the virus being present in human cancer has been reported although reports for it being present in ], some nonHodgkin's ] and other human cancers have been published. This is confounded by the high level of cross-reactivity for SV40 with known human polyomaviruses (BK virus and JC virus) that are widespread and by common use of SV40 DNA as a near universal reagent in scientific laboratories.<ref name=pmid14566815/> Most virologists dismiss SV40 as a cause for human cancers.<ref name=pmid16963733/> | |||
| ===Possible association with prostate cancer=== | |||
| BK viral genomes have been found in benign and malignant prostate tissue.<ref name=Delbue2013>Delbue S, Matei DV, Carloni C, Pecchenini V, Carluccio S, Villani S, Tringali V, Brescia A, Ferrante P (2013) Evidence supporting the association of polyomavirus BK genome with prostate cancer. Med Microbiol Immunol </ref> It has been suggested that this virus may play a role in the development of malignancy but further work in this area is required. | |||
| == Diagnosis == | |||
| The diagnosis of polyomavirus almost always occurs after the primary infection as it is either asymptomatic or sub-clinical. Antibody assays are commonly used to detect presence of antibodies against individual viruses.<ref name="Cinthia B. Drachenberg 2005">{{cite journal |author=Drachenberg CB, Hirsch HH, Ramos E, Papadimitriou JC |title=Polyomavirus disease in renal transplantation: review of pathological findings and diagnostic methods |journal=Human Pathology |volume=36 |issue=12 |pages=1245–55 |year=2005 |month=December |pmid=16311117 |doi=10.1016/j.humpath.2005.08.009}}</ref> Competition assays are frequently needed to distinguish among highly similar polyomaviruses.<ref>{{cite journal |author=Viscidi RP, Clayman B |title=Serological cross reactivity between polyomavirus capsids |journal=Advances in Experimental Medicine and Biology |volume=577 |issue= |pages=73–84 |year=2006 |pmid=16626028 |doi=10.1007/0-387-32957-9_5 |series=Advances in Experimental Medicine and Biology |isbn=978-0-387-29233-5}}</ref> | |||
| In cases of progressive multifocal leucoencephalopathy (PML), a cross-reactive antibody to SV40 T antigen (commonly Pab419) is used to stain tissues directly for the presence of JC virus T antigen. PCR can be used on a biopsy of the tissue or ] to amplify the polyomavirus DNA. This allows not only the detection of polyomavirus but also which sub type it is.<ref>{{cite journal |author=Drews K, Bashir T, Dörries K |title=Quantification of human polyomavirus JC in brain tissue and cerebrospinal fluid of patients with progressive multifocal leukoencephalopathy by competitive PCR |journal=Journal of Virological Methods |volume=84 |issue=1 |pages=23–36 |year=2000 |month=January |pmid=10644084 |doi=10.1016/S0166-0934(99)00128-7}}</ref> | |||
| There are three main diagnostic techniques used for the diagnosis of the reactivation of polyomavirus in polyomavirus nephropathy (PVN): urine cytology, quantification of the viral load in both urine and blood, and a ].<ref name="Cinthia B. Drachenberg 2005"/> | There are three main diagnostic techniques used for the diagnosis of the reactivation of polyomavirus in polyomavirus nephropathy (PVN): urine cytology, quantification of the viral load in both urine and blood, and a ].<ref name="Cinthia B. Drachenberg 2005"/> | ||
| The reactivation of polyomavirus in the kidneys and urinary tract causes the shedding of infected cells, virions, and/or viral proteins in the urine. This allows urine cytology to examine these cells, which if there is polyomavirus inclusion of the nucleus, is diagnostic of infection.<ref>{{cite journal | |
The reactivation of polyomavirus in the kidneys and urinary tract causes the shedding of infected cells, virions, and/or viral proteins in the urine. This allows urine cytology to examine these cells, which if there is polyomavirus inclusion of the nucleus, is diagnostic of infection.<ref>{{cite journal | vauthors = Nickeleit V, Hirsch HH, Binet IF, Gudat F, Prince O, Dalquen P, Thiel G, Mihatsch MJ | title = Polyomavirus infection of renal allograft recipients: from latent infection to manifest disease | journal = Journal of the American Society of Nephrology | volume = 10 | issue = 5 | pages = 1080–9 | date = May 1999 | doi = 10.1681/ASN.V1051080 | pmid = 10232695 | url = http://jasn.asnjournals.org/cgi/pmidlookup?view=long&pmid=10232695 | doi-access = free }}</ref> Also as the urine of an infected individual will contain virions and/or viral DNA, quantitation of the viral load can be done through PCR.<ref>{{cite journal | vauthors = Randhawa PS, Vats A, Zygmunt D, Swalsky P, Scantlebury V, Shapiro R, Finkelstein S | title = Quantitation of viral DNA in renal allograft tissue from patients with BK virus nephropathy | journal = Transplantation | volume = 74 | issue = 4 | pages = 485–8 | date = August 2002 | pmid = 12352906 | doi = 10.1097/00007890-200208270-00009 | s2cid = 30574884 | doi-access = free }}</ref> This is also true for the blood. | ||
| Renal biopsy can also be used if the two methods just described are inconclusive or if the specific viral load for the renal tissue is desired. Similarly to the urine cytology, the renal cells are examined under light microscopy for polyomavirus inclusion of the nucleus, as well as cell lysis and viral partials in the extra cellular fluid. The viral load as before is also measure by PCR. | Renal biopsy can also be used if the two methods just described are inconclusive or if the specific viral load for the renal tissue is desired. Similarly to the urine cytology, the renal cells are examined under light microscopy for polyomavirus inclusion of the nucleus, as well as cell lysis and viral partials in the extra cellular fluid. The viral load as before is also measure by PCR.{{citation needed|date=February 2015}} | ||
| Tissue staining using a monoclonal antibody against MCV T antigen shows utility in differentiating Merkel cell carcinoma from other small, round cell tumors.<ref>{{cite journal | vauthors = Busam KJ, Jungbluth AA, Rekthman N, Coit D, Pulitzer M, Bini J, Arora R, Hanson NC, Tassello JA, Frosina D, Moore P, Chang Y | title = Merkel cell polyomavirus expression in merkel cell carcinomas and its absence in combined tumors and pulmonary neuroendocrine carcinomas | journal = The American Journal of Surgical Pathology | volume = 33 | issue = 9 | pages = 1378–85 | date = September 2009 | pmid = 19609205 | pmc = 2932664 | doi = 10.1097/PAS.0b013e3181aa30a5 }}</ref> Blood tests to detect MCV antibodies have been developed and show that infection with the virus is widespread although Merkel cell carcinoma patients have exceptionally higher antibody responses than asymptomatically infected persons.<ref name="kean"/><ref>{{cite journal | vauthors = Tolstov YL, Pastrana DV, Feng H, Becker JC, Jenkins FJ, Moschos S, Chang Y, Buck CB, Moore PS | title = Human Merkel cell polyomavirus infection II. MCV is a common human infection that can be detected by conformational capsid epitope immunoassays | journal = International Journal of Cancer | volume = 125 | issue = 6 | pages = 1250–6 | date = September 2009 | pmid = 19499548 | pmc = 2747737 | doi = 10.1002/ijc.24509 }}</ref><ref>{{cite journal | vauthors = Pastrana DV, Tolstov YL, Becker JC, Moore PS, Chang Y, Buck CB | title = Quantitation of human seroresponsiveness to Merkel cell polyomavirus | journal = PLOS Pathogens | volume = 5 | issue = 9 | pages = e1000578 | date = September 2009 | pmid = 19750217 | pmc = 2734180 | doi = 10.1371/journal.ppat.1000578 | doi-access = free }}</ref><ref>{{cite journal | vauthors = Carter JJ, Paulson KG, Wipf GC, Miranda D, Madeleine MM, Johnson LG, Lemos BD, Lee S, Warcola AH, Iyer JG, Nghiem P, Galloway DA | title = Association of Merkel cell polyomavirus-specific antibodies with Merkel cell carcinoma | journal = Journal of the National Cancer Institute | volume = 101 | issue = 21 | pages = 1510–22 | date = November 2009 | pmid = 19776382 | pmc = 2773184 | doi = 10.1093/jnci/djp332 }}</ref> | |||
| Tissue staining using a monoclonal antibody against MCV T antigen shows utility in differentiating Merkel cell carcinoma from other small, round cell tumors.<ref>{{cite journal | |||
| |last=Busam |first=Klaus J. |last2=Jungbluth |first2=Achim A. |last3=Rekthman |first3=Natasha |coauthors=and nine additional authors |year=2009 |month=September |title=Merkel Cell Polyomavirus Expression in Merkel Cell Carcinomas and Its Absence in Combined Tumors and Pulmonary Neuroendocrine Carcinomas |journal=] |volume=33 |issue=9 |pages=1378–1385 |doi=10.1097/PAS.0b013e3181aa30a5 |url=http://journals.lww.com/ajsp/Abstract/2009/09000/Merkel_Cell_Polyomavirus_Expression_in_Merkel_Cell.15.aspx |accessdate=March 3, 2013 }}{{subscription required}}</ref> Blood tests to detect MCV antibodies have been developed and show that infection with the virus is widespread although Merkel cell carcinoma patients have exceptionally higher antibody responses than asymptomatically infected persons.<ref>{{cite journal |author=Kean JM, Rao S, Wang M, Garcea RL |editor1-last=Atwood |editor1-first=Walter J. |title=Seroepidemiology of human polyomaviruses |journal=PLoS Pathogens |volume=5 |issue=3 |pages=e1000363 |year=2009 |month=March |pmid=19325891 |pmc=2655709 |doi=10.1371/journal.ppat.1000363}}</ref><ref>{{cite journal |author=Tolstov YL, Pastrana DV, Feng H, ''et al.'' |title=Human Merkel cell polyomavirus infection II. MCV is a common human infection that can be detected by conformational capsid epitope immunoassays |journal=International Journal of Cancer |volume=125 |issue=6 |pages=1250–6 |year=2009 |month=September |pmid=19499548 |pmc=2747737 |doi=10.1002/ijc.24509}}</ref><ref>{{cite journal |author=Pastrana DV, Tolstov YL, Becker JC, Moore PS, Chang Y, Buck CB |editor1-last=Garcea |editor1-first=Robert L. |title=Quantitation of human seroresponsiveness to Merkel cell polyomavirus |journal=PLoS Pathogens |volume=5 |issue=9 |pages=e1000578 |year=2009 |month=September |pmid=19750217 |pmc=2734180 |doi=10.1371/journal.ppat.1000578}}</ref><ref>{{cite journal |author=Carter JJ, Paulson KG, Wipf GC, ''et al.'' |title=Association of merkel cell polyomavirus-specific antibodies with merkel cell carcinoma |journal=Journal of the National Cancer Institute |volume=101 |issue=21 |pages=1510–22 |year=2009 |month=November |pmid=19776382 |doi=10.1093/jnci/djp332 |pmc=2773184}}</ref> | |||
| === Use in tracing human migration === | |||
| These viruses have been found in ] tumours.<ref name=Hachana2011>Hachana M, Amara K, Ziadi S, Gacem RB, Korbi S, Trimeche M (2011) Investigation of human JC and BK polyomaviruses in breast carcinomas. Breast Cancer Res Treat</ref> The importance of this finding—if any—is not known. | |||
| The JC virus offers a promising genetic marker for human evolution and migration.<ref>{{cite book |author1=Elizabeth Matisoo-Smith |author2=K. Ann Horsburgh |title=DNA for Archaeologists |publisher=Routledge |date=2012 |isbn=978-1598746815}}</ref><ref name= Sugimoto_1997>{{cite journal | vauthors = Sugimoto C, Kitamura T, Guo J, Al-Ahdal MN, Shchelkunov SN, Otova B, Ondrejka P, Chollet JY, El-Safi S, Ettayebi M, Grésenguet G, Kocagöz T, Chaiyarasamee S, Thant KZ, Thein S, Moe K, Kobayashi N, Taguchi F, Yogo Y |title= Typing of urinary JC virus DNA offers a novel means of tracing human migrations |journal= Proc Natl Acad Sci U S A |date=August 19, 1997 | volume = 94| issue =17 |pages=9191–9196|doi=10.1073/pnas.94.17.9191 | |||
| |pmid=9256458|pmc=23108|doi-access = free|bibcode= 1997PNAS...94.9191S }}</ref> It is carried by 70–90 percent of humans and is usually transmitted from parents to offspring. This method does not appear to be reliable for tracing the ].{{citation needed|date=November 2022}} | |||
| == |
== History == | ||
| ] was the first polyomavirus discovered, having been reported by ] in 1953 as an extract of mouse ]s capable of inducing ] tumors.<ref>{{cite journal | vauthors = Gross L | title = A filterable agent, recovered from Ak leukemic extracts, causing salivary gland carcinomas in C3H mice | journal = Proceedings of the Society for Experimental Biology and Medicine | volume = 83 | issue = 2 | pages = 414–21 | date = June 1953 | pmid = 13064287 | doi = 10.3181/00379727-83-20376 | s2cid = 34223353 }}</ref> The causative agent was identified as a virus by ] and ], after whom it was once called "SE polyoma".<ref>{{cite journal | vauthors = Stewart SE, Eddy BE, Borgese N | title = Neoplasms in mice inoculated with a tumor agent carried in tissue culture | journal = Journal of the National Cancer Institute | volume = 20 | issue = 6 | pages = 1223–43 | date = June 1958 | pmid = 13549981 | doi=10.1093/jnci/20.6.1223}}</ref><ref>{{cite journal | vauthors = Eddy BE, Stewart SE | title = Characteristics of the SE polyoma virus | journal = American Journal of Public Health and the Nation's Health | volume = 49 | issue = 11 | pages = 1486–92 | date = November 1959 | pmid = 13819251 | pmc = 1373056 | doi = 10.2105/AJPH.49.11.1486 }}</ref><ref name=pathology>{{cite journal | vauthors = Schowalter RM, Pastrana DV, Buck CB | title = Glycosaminoglycans and sialylated glycans sequentially facilitate Merkel cell polyomavirus infectious entry | journal = PLOS Pathogens | volume = 7 | issue = 7 | pages = e1002161 | date = July 2011 | pmid = 21829355 | doi = 10.1371/journal.ppat.1002161 | pmc=3145800 | doi-access = free }}</ref> The term "polyoma" refers to the viruses' ability to produce multiple (poly-) tumors (-oma) under certain conditions. The name has been criticized as a "meatless linguistic sandwich" ("meatless" because both ]s in "polyoma" are affixes) giving little insight into the viruses' biology; in fact, subsequent research has found that most polyomaviruses rarely cause clinically significant disease in their host organisms under natural conditions.<ref name="gottlieb">{{cite journal | vauthors = Gottlieb KA, Villarreal LP | title = Natural biology of polyomavirus middle T antigen | journal = Microbiology and Molecular Biology Reviews | volume = 65 | issue = 2 | pages = 288–318; second and third pages, table of contents | date = June 2001 | pmid = 11381103 | pmc = 99028 | doi = 10.1128/mmbr.65.2.288-318.2001 }}</ref> | |||
| Dozens of polyomaviruses have been identified and sequenced as of 2017, infecting mainly birds and mammals. Two polyomaviruses are known to infect fish, the ]<ref name="peretti_2015">{{cite journal | vauthors = Peretti A, FitzGerald PC, Bliskovsky V, Pastrana DV, Buck CB | title = Genome Sequence of a Fish-Associated Polyomavirus, Black Sea Bass (Centropristis striata) Polyomavirus 1 | journal = Genome Announcements | volume = 3 | issue = 1 | pages = e01476-14 | date = January 2015 | pmid = 25635011 | pmc = 4319505 | doi = 10.1128/genomeA.01476-14 }}</ref> and ].<ref name="lopez_2016">{{cite journal | vauthors = López-Bueno A, Mavian C, Labella AM, Castro D, Borrego JJ, Alcami A, Alejo A | title = Concurrence of Iridovirus, Polyomavirus, and a Unique Member of a New Group of Fish Papillomaviruses in Lymphocystis Disease-Affected Gilthead Sea Bream | journal = Journal of Virology | volume = 90 | issue = 19 | pages = 8768–79 | date = October 2016 | pmid = 27440877 | pmc = 5021401 | doi = 10.1128/JVI.01369-16 }}</ref> A total of fourteen polyomaviruses are known to infect humans.<ref name=Polyomaviridae2016 /> | |||
| There is no known treatment for infection with these viruses. However it appears that some of ]s may have therapeutic potential.<ref>Sharma BN, Li R, Bernhoff E, Gutteberg TJ, Rinaldo CH (2011) Fluoroquinolones inhibit human polyomavirus BK (BKV) replication in primary human kidney cells. Antiviral Res</ref> | |||
| == References == | == References == | ||
| {{Reflist| |
{{Reflist|30em}} | ||
| == External links == | == External links == | ||
| * | |||
| * | * | ||
| * | |||
| {{Baltimore classification}} | {{Baltimore classification}} | ||
| {{Taxonbar|from=Q18670504}} | |||
| {{Authority control}} | |||
| ] | ] | ||
| ] | |||
| ] | |||
Latest revision as of 00:50, 31 December 2024
Family of viruses
| Polyomaviridae | |
|---|---|
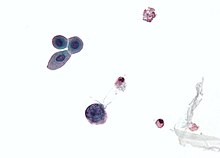
| |
| Micrograph showing a polyomavirus infected cell—large (blue) cell below-center-left. Urine cytology specimen. | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Monodnaviria |
| Kingdom: | Shotokuvirae |
| Phylum: | Cossaviricota |
| Class: | Papovaviricetes |
| Order: | Sepolyvirales |
| Family: | Polyomaviridae |
| Genera | |
Polyomaviridae is a family of viruses whose natural hosts are primarily mammals and birds. As of 2024, there are eight recognized genera. Fourteen species are known to infect humans, while others, such as Simian Virus 40, have been identified in humans to a lesser extent. Most of these viruses are very common and typically asymptomatic in most human populations studied. BK virus is associated with nephropathy in renal transplant and non-renal solid organ transplant patients, JC virus with progressive multifocal leukoencephalopathy, and Merkel cell virus with Merkel cell cancer.
Structure and genome

Polyomaviruses are non-enveloped double-stranded DNA viruses with circular genomes of around 5000 base pairs. With such a small size, they are ranked among the smallest known double stranded DNA viruses. The genome is packaged in a viral capsid of about 40-50 nanometers in diameter, which is icosahedral in shape (T=7 symmetry). The capsid is composed of 72 pentameric capsomeres of a protein called VP1, which is capable of self-assembly into a closed icosahedron; each pentamer of VP1 is associated with one molecule of one of the other two capsid proteins, VP2 or VP3.

The genome of a typical polyomavirus codes for between five and nine proteins, divided into two transcriptional regions called the early and late regions due to the time during infection in which they are transcribed. Each region is transcribed by the host cell's RNA polymerase II as a single pre-messenger RNA containing multiple genes. The early region usually codes for two proteins, the small and large tumor antigens, produced by alternative splicing. The late region contains the three capsid structural proteins VP1, VP2, and VP3, produced by alternative translational start sites. Additional genes and other variations on this theme are present in some viruses: for example, rodent polyomaviruses have a third protein called middle tumor antigen in the early region, which is extremely efficient at inducing cellular transformation; SV40 has an additional capsid protein VP4; some examples have an additional regulatory protein called agnoprotein expressed from the late region. The genome also contains a non-coding control or regulatory region containing the early and late regions' promoters, transcriptional start sites, and the origin of replication.
Replication and life cycle
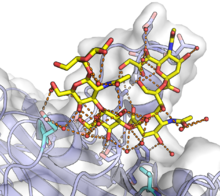
The polyomavirus life cycle begins with entry into a host cell. Cellular receptors for polyomaviruses are sialic acid residues of glycans, commonly gangliosides. The attachment of polyomaviruses to host cells is mediated by the binding of VP1 to sialylated glycans on the cell surface. In some particular viruses, additional cell-surface interactions occur; for example, the JC virus is believed to require interaction with the 5HT2A receptor and the Merkel cell virus with heparan sulfate. However, in general virus-cell interactions are mediated by commonly occurring molecules on the cell surface, and therefore are likely not a major contributor to individual viruses' observed cell-type tropism. After binding to molecules on the cell surface, the virion is endocytosed and enters the endoplasmic reticulum - a behavior unique among known non-enveloped viruses - where the viral capsid structure is likely to be disrupted by action of host cell disulfide isomerase enzymes.
The details of transit to the nucleus are not clear and may vary among individual polyomaviruses. It has been frequently reported that an intact, albeit distorted, virion particle is released from the endoplasmic reticulum into the cell cytoplasm, where the genome is released from the capsid, possibly due to the low calcium concentration in the cytoplasm. Both expression of viral genes and replication of the viral genome occur in the nucleus using host cell machinery. The early genes - comprising at minimum the small tumor antigen (ST) and large tumor antigen (LT) - are expressed first, from a single alternatively spliced messenger RNA strand. These proteins serve to manipulate the host's cell cycle - dysregulating the transition from G1 phase to S phase, when the host cell's genome is replicated - because host cell DNA replication machinery is needed for viral genome replication. The precise mechanism of this dysregulation depends on the virus; for example, SV40 LT can directly bind host cell p53, but murine polyomavirus LT does not. LT induces DNA replication from the viral genome's non-coding control region (NCCR), after which expression of the early mRNA is reduced and expression of the late mRNA, which encodes the viral capsid proteins, begins. As these interactions begin, the LTs belonging to several polyomaviruses, including Merkel cell polyomavirus, present oncogenic potential. Several mechanisms have been described for regulating the transition from early to late gene expression, including the involvement of the LT protein in repressing the early promoter, the expression of un-terminated late mRNAs with extensions complementary to early mRNA, and the expression of regulatory microRNA. Expression of the late genes results in accumulation of the viral capsid proteins in the host cell cytoplasm. Capsid components enter the nucleus in order to encapsidate new viral genomic DNA. New virions may be assembled in viral factories. The mechanism of viral release from the host cell varies among polyomaviruses; some express proteins that facilitate cell exit, such as the agnoprotein or VP4. In some cases high levels of encapsidated virus result in cell lysis, releasing the virions.
Viral proteins
Tumor antigens
The large tumor antigen plays a key role in regulating the viral life cycle by binding to the viral origin of DNA replication where it promotes DNA synthesis. Also as the polyomavirus relies on the host cell machinery to replicate the host cell needs to be in s-phase for this to begin. Due to this, large T-antigen also modulates cellular signaling pathways to stimulate progression of the cell cycle by binding to a number of cellular control proteins. This is achieved by a two prong attack of inhibiting tumor suppressing genes p53 and members of the retinoblastoma (pRB) family, and stimulating cell growth pathways by binding cellular DNA, ATPase-helicase, DNA polymerase α association, and binding of transcription preinitiation complex factors. This abnormal stimulation of the cell cycle is a powerful force for oncogenic transformation.
The small tumor antigen protein is also able to activate several cellular pathways that stimulate cell proliferation. Polyomavirus small T antigens commonly target protein phosphatase 2A (PP2A), a key multisubunit regulator of multiple pathways including Akt, the mitogen-activated protein kinase (MAPK) pathway, and the stress-activated protein kinase (SAPK) pathway. Merkel cell polyomavirus small T antigen encodes a unique domain, called the LT-stabilization domain (LSD), that binds to and inhibits the FBXW7 E3 ligase regulating both cellular and viral oncoproteins. Unlike for SV40, the MCV small T antigen directly transforms rodent cells in vitro.
The middle tumor antigen is used in model organisms developed to study cancer, such as the MMTV-PyMT system where middle T is coupled to the MMTV promoter. There it functions as an oncogene, while the tissue where the tumor develops is determined by the MMTV promoter.
Capsid proteins
The polyomavirus capsid consists of one major component, major capsid protein VP1, and one or two minor components, minor capsid proteins VP2 and VP3. VP1 pentamers form the closed icosahedral viral capsid, and in the interior of the capsid each pentamer is associated with one molecule of either VP2 or VP3. Some polyomaviruses, such as Merkel cell polyomavirus, do not encode or express VP3. The capsid proteins are expressed from the late region of the genome.
Agnoprotein
The agnoprotein is a small multifunctional phospho-protein found in the late coding part of the genome of some polyomaviruses, most notably BK virus, JC virus, and SV40. It is essential for proliferation in the viruses that express it and is thought to be involved in regulating the viral life cycle, particularly replication and viral exit from the host cell, but the exact mechanisms are unclear.
Taxonomy
The polyomaviruses are members of group I (dsDNA viruses). The classification of polyomaviruses has been the subject of several proposed revisions as new members of the group are discovered. Formerly, polyomaviruses and papillomaviruses, which share many structural features but have very different genomic organizations, were classified together in the now-obsolete family Papovaviridae. (The name Papovaviridae derived from three abbreviations: Pa for Papillomavirus, Po for Polyomavirus, and Va for "vacuolating.") The polyomaviruses were divided into three major clades (that is, genetically related groups): the SV40 clade, the avian clade, and the murine polyomavirus clade. A subsequent proposed reclassification by the International Committee on Taxonomy of Viruses (ICTV) recommended dividing the family of Polyomaviridae into three genera:
- Genus Orthopolyomavirus (type species SV40)
- Genus Wukipolyomavirus (type species KI polyomavirus)
- Genus Avipolyomavirus (type species Avian polyomavirus)
The current ICTV classification system recognises six genera and 117 species, of which five could not be assigned a genus. This system retains the distinction between avian and mammalian viruses, grouping the avian subset into the genus Gammapolyomavirus. The six genera are:
- Alphapolyomavirus
- Betapolyomavirus
- Deltapolyomavirus
- Epsilonpolyomavirus
- Gammapolyomavirus
- Zetapolyomavirus
The following species are unassigned to a genus:
- Centropristis striata polyomavirus 1
- Rhynchobatus djiddensis polyomavirus 1
- Sparus aurata polyomavirus 1
- Trematomus bernacchii polyomavirus 1
- Trematomus pennellii polyomavirus 1
Description of additional viruses is ongoing. These include the sea otter polyomavirus 1 and Alpaca polyomavirus Another virus is the giant panda polyomavirus 1. Another virus has been described from sigmodontine rodents. Another - tree shrew polyomavirus 1 - has been described in the tree shrew.
Human polyomaviruses
Most polyomaviruses do not infect humans. Of the polyomaviruses cataloged as of 2017, a total of 14 were known with human hosts. However, some polyomaviruses are associated with human disease, particularly in immunocompromised individuals. MCV is highly divergent from the other human polyomaviruses and is most closely related to murine polyomavirus. Trichodysplasia spinulosa-associated polyomavirus (TSV) is distantly related to MCV. Two viruses—HPyV6 and HPyV7—are most closely related to KI and WU viruses, while HPyV9 is most closely related to the African green monkey-derived lymphotropic polyomavirus (LPV).
A fourteenth virus has been described. Lyon IARC polyomavirus is related to raccoon polyomavirus.
List of human polyomaviruses
The following 14 polyomaviruses with human hosts had been identified and had their genomes sequenced as of 2017:
| Species | Proposed genus | Virus name | Abbreviation | NCBI RefSeq | Year of discovery | Clinical correlate (if any) | References |
|---|---|---|---|---|---|---|---|
| Human polyomavirus 5 | Alpha | Merkel cell polyomavirus | MCPyV | NC_010277 | 2008 | Merkel cell cancer | |
| Human polyomavirus 8 | Alpha | Trichodysplasia spinulosa polyomavirus | TSPyV | NC_014361 | 2010 | Trichodysplasia spinulosa | |
| Human polyomavirus 9 | Alpha | Human polyomavirus 9 | HPyV9 | NC_015150 | 2011 | None known | |
| Human polyomavirus 12 | Alpha | Human polyomavirus 12 | HPyV12 | NC_020890 | 2013 | None known | |
| Human polyomavirus 13 | Alpha | New Jersey polyomavirus | NJPyV | NC_024118 | 2014 | None known | |
| Human polyomavirus 1 | Beta | BK polyomavirus | BKPyV | NC_001538 | 1971 | Polyomavirus-associated nephropathy; haemorrhagic cystitis | |
| Human polyomavirus 2 | Beta | JC polyomavirus | JCPyV | NC_001699 | 1971 | Progressive multifocal leukoencephalopathy | |
| Human polyomavirus 3 | Beta | KI polyomavirus | KIPyV | NC_009238 | 2007 | None known | |
| Human polyomavirus 4 | Beta | WU polyomavirus | WUPyV | NC_009539 | 2007 | None known | |
| Human polyomavirus 6 | Delta | Human polyomavirus 6 | HPyV6 | NC_014406 | 2010 | HPyV6 associated pruritic and dyskeratotic dermatosis (H6PD) | |
| Human polyomavirus 7 | Delta | Human polyomavirus 7 | HPyV7 | NC_014407 | 2010 | HPyV7-related epithelial hyperplasia | |
| Human polyomavirus 10 | Delta | MW polyomavirus | MWPyV | NC_018102 | 2012 | None known | |
| Human polyomavirus 11 | Delta | STL polyomavirus | STLPyV | NC_020106 | 2013 | None known | |
| Human polyomavirus 14 | Alpha | Lyon IARC polyomavirus | LIPyV | NC_034253.1 | 2017 | None known |
Deltapolyomavirus contains only the four human viruses shown in the above table. The Alpha and Beta groups contain viruses that infect a variety of mammals. The Gamma group contains the avian viruses. Clinically significant disease associations are shown only where causality is expected.
Antibodies to the monkey lymphotropic polyomavirus have been detected in humans suggesting that this virus - or a closely related virus - can infect humans.
Clinical relevance
All the polyomaviruses are highly common childhood and young adult infections. Most of these infections appear to cause little or no symptoms. These viruses are probably lifelong persistent among almost all adults. Diseases caused by human polyomavirus infections are most common among immunocompromised people; disease associations include BK virus with nephropathy in renal transplant and non-renal solid organ transplant patients, JC virus with progressive multifocal leukoencephalopathy, and Merkel cell virus (MCV) with Merkel cell cancer.
SV40
Main article: SV40SV40 replicates in the kidneys of monkeys without causing disease, but can cause cancer in rodents under laboratory conditions. In the 1950s and early 1960s, well over 100 million people may have been exposed to SV40 due to previously undetected SV40 contamination of polio vaccine, prompting concern about the possibility that the virus might cause disease in humans. Although it has been reported as present in some human cancers, including brain tumors, bone tumors, mesotheliomas, and non-Hodgkin's lymphomas, accurate detection is often confounded by high levels of cross-reactivity for SV40 with widespread human polyomaviruses. Most virologists dismiss SV40 as a cause for human cancers.
Diagnosis
The diagnosis of polyomavirus almost always occurs after the primary infection as it is either asymptomatic or sub-clinical. Antibody assays are commonly used to detect presence of antibodies against individual viruses. Competition assays are frequently needed to distinguish among highly similar polyomaviruses.
In cases of progressive multifocal leucoencephalopathy (PML), a cross-reactive antibody to SV40 T antigen (commonly Pab419) is used to stain tissues directly for the presence of JC virus T antigen. PCR can be used on a biopsy of the tissue or cerebrospinal fluid to amplify the polyomavirus DNA. This allows not only the detection of polyomavirus but also which sub type it is.
There are three main diagnostic techniques used for the diagnosis of the reactivation of polyomavirus in polyomavirus nephropathy (PVN): urine cytology, quantification of the viral load in both urine and blood, and a renal biopsy. The reactivation of polyomavirus in the kidneys and urinary tract causes the shedding of infected cells, virions, and/or viral proteins in the urine. This allows urine cytology to examine these cells, which if there is polyomavirus inclusion of the nucleus, is diagnostic of infection. Also as the urine of an infected individual will contain virions and/or viral DNA, quantitation of the viral load can be done through PCR. This is also true for the blood.
Renal biopsy can also be used if the two methods just described are inconclusive or if the specific viral load for the renal tissue is desired. Similarly to the urine cytology, the renal cells are examined under light microscopy for polyomavirus inclusion of the nucleus, as well as cell lysis and viral partials in the extra cellular fluid. The viral load as before is also measure by PCR.
Tissue staining using a monoclonal antibody against MCV T antigen shows utility in differentiating Merkel cell carcinoma from other small, round cell tumors. Blood tests to detect MCV antibodies have been developed and show that infection with the virus is widespread although Merkel cell carcinoma patients have exceptionally higher antibody responses than asymptomatically infected persons.
Use in tracing human migration
The JC virus offers a promising genetic marker for human evolution and migration. It is carried by 70–90 percent of humans and is usually transmitted from parents to offspring. This method does not appear to be reliable for tracing the recent African origin of modern humans.
History
Murine polyomavirus was the first polyomavirus discovered, having been reported by Ludwik Gross in 1953 as an extract of mouse leukemias capable of inducing parotid gland tumors. The causative agent was identified as a virus by Sarah Stewart and Bernice Eddy, after whom it was once called "SE polyoma". The term "polyoma" refers to the viruses' ability to produce multiple (poly-) tumors (-oma) under certain conditions. The name has been criticized as a "meatless linguistic sandwich" ("meatless" because both morphemes in "polyoma" are affixes) giving little insight into the viruses' biology; in fact, subsequent research has found that most polyomaviruses rarely cause clinically significant disease in their host organisms under natural conditions.
Dozens of polyomaviruses have been identified and sequenced as of 2017, infecting mainly birds and mammals. Two polyomaviruses are known to infect fish, the black sea bass and gilthead seabream. A total of fourteen polyomaviruses are known to infect humans.
References
- Moens U, Calvignac-Spencer S, Lauber C, Ramqvist T, Feltkamp MC, Daugherty MD, Verschoor EJ, Ehlers B (June 2017). "ICTV Virus Taxonomy Profile: Polyomaviridae". The Journal of General Virology. 98 (6): 1159–1160. doi:10.1099/jgv.0.000839. PMC 5656788. PMID 28640744.
- ^ "ICTV Report Polyomaviridae".
- taxonomy. "Taxonomy browser (Polyomaviridae)". www.ncbi.nlm.nih.gov. Retrieved 19 March 2024.
- ^ Calvignac-Spencer S, Feltkamp MC, Daugherty MD, Moens U, Ramqvist T, Johne R, Ehlers B (June 2016). "A taxonomy update for the family Polyomaviridae". Archives of Virology. 161 (6): 1739–50. doi:10.1007/s00705-016-2794-y. hdl:10037/13151. PMID 26923930.
- ^ DeCaprio JA, Garcea RL (April 2013). "A cornucopia of human polyomaviruses". Nature Reviews. Microbiology. 11 (4): 264–76. doi:10.1038/nrmicro2992. PMC 3928796. PMID 23474680.
- Gossai A, Waterboer T, Nelson HH, Michel A, Willhauck-Fleckenstein M, Farzan SF, Hoen AG, Christensen BC, Kelsey KT, Marsit CJ, Pawlita M, Karagas MR (January 2016). "Seroepidemiology of Human Polyomaviruses in a US Population". American Journal of Epidemiology. 183 (1): 61–9. doi:10.1093/aje/kwv155. PMC 5006224. PMID 26667254.
- ^ Kean JM, Rao S, Wang M, Garcea RL (March 2009). "Seroepidemiology of human polyomaviruses". PLOS Pathogens. 5 (3): e1000363. doi:10.1371/journal.ppat.1000363. PMC 2655709. PMID 19325891.
- ^ Jamboti JS (August 2016). "BK virus nephropathy in renal transplant recipients". Nephrology. 21 (8): 647–54. doi:10.1111/nep.12728. PMID 26780694.
- ^ Kuppachi S, Kaur D, Holanda DG, Thomas CP (April 2016). "BK polyoma virus infection and renal disease in non-renal solid organ transplantation". Clinical Kidney Journal. 9 (2): 310–8. doi:10.1093/ckj/sfv143. PMC 4792618. PMID 26985385.
- ^ Adang L, Berger J (2015). "Progressive Multifocal Leukoencephalopathy". F1000Research. 4: 1424. doi:10.12688/f1000research.7071.1. PMC 4754031. PMID 26918152.
- ^ Feng H, Shuda M, Chang Y, Moore PS (February 2008). "Clonal integration of a polyomavirus in human Merkel cell carcinoma". Science. 319 (5866): 1096–100. Bibcode:2008Sci...319.1096F. doi:10.1126/science.1152586. PMC 2740911. PMID 18202256.
- Shackelton LA, Rambaut A, Pybus OG, Holmes EC (October 2006). "JC virus evolution and its association with human populations". J Virol. 80 (20): 9928–9933. doi:10.1128/JVI.00441-06. PMC 1617318. PMID 17005670.
- ^ "Viral Zone". ExPASy. Retrieved 15 June 2015.
- Salunke DM, Caspar DL, Garcea RL (September 1986). "Self-assembly of purified polyomavirus capsid protein VP1". Cell. 46 (6): 895–904. doi:10.1016/0092-8674(86)90071-1. PMID 3019556. S2CID 25800023.
- ^ Gaynor AM, Nissen MD, Whiley DM, Mackay IM, Lambert SB, Wu G, Brennan DC, Storch GA, Sloots TP, Wang D (May 2007). "Identification of a novel polyomavirus from patients with acute respiratory tract infections". PLOS Pathogens. 3 (5): e64. doi:10.1371/journal.ppat.0030064. PMC 1864993. PMID 17480120.
- ^ International Agency for Research on Cancer (2013). "Introduction to Polyomaviruses" (PDF). IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. 104: 121–131.
- ^ Buch MH, Liaci AM, O'Hara SD, Garcea RL, Neu U, Stehle T (October 2015). "Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity". PLOS Pathogens. 11 (10): e1005104. doi:10.1371/journal.ppat.1005104. PMC 4608799. PMID 26474293.
- Schowalter RM, Pastrana DV, Buck CB (July 2011). "Glycosaminoglycans and sialylated glycans sequentially facilitate Merkel cell polyomavirus infectious entry". PLOS Pathogens. 7 (7): e1002161. doi:10.1371/journal.ppat.1002161. PMC 3145800. PMID 21829355.
- ^ Inoue T, Tsai B (January 2013). "How viruses use the endoplasmic reticulum for entry, replication, and assembly". Cold Spring Harbor Perspectives in Biology. 5 (1): a013250. doi:10.1101/cshperspect.a013250. PMC 3579393. PMID 23284050.
- ^ Gjoerup O, Chang Y (2010). "Update on human polyomaviruses and cancer". Advances in Cancer Research. 106: 1–51. doi:10.1016/S0065-230X(10)06001-X. ISBN 9780123747716. PMID 20399955.
- Andrabi S, Hwang JH, Choe JK, Roberts TM, Schaffhausen BS (2011). "Comparisons between Murine Polyomavirus and Simian Virus 40 Show Significant Differences in Small T Antigen Function". Journal of Virology. 85 (20): 10649–10658. doi:10.1128/JVI.05034-11. PMC 3187521. PMID 21835797.
- Rotondo JC, Bononi I, Puozzo A, Govoni M, Foschi V, Lanza G, Gafà R, Gaboriaud P, Touzé FA, Selvatici R, Martini F, Tognon M (July 2017). "Merkel Cell Carcinomas Arising in Autoimmune Disease Affected Patients Treated with Biologic Drugs, Including Anti-TNF". Clinical Cancer Research. 23 (14): 3929–3934. doi:10.1158/1078-0432.CCR-16-2899. hdl:11392/2378829. PMID 28174236.
- White MK, Gordon J, Reiss K, Del Valle L, Croul S, Giordano A, Darbinyan A, Khalili K (December 2005). "Human polyomaviruses and brain tumors". Brain Research. Brain Research Reviews. 50 (1): 69–85. doi:10.1016/j.brainresrev.2005.04.007. PMID 15982744. S2CID 20990837.
- Kazem S, van der Meijden E, Wang RC, Rosenberg AS, Pope E, Benoit T, Fleckman P, Feltkamp MC (2014). "Polyomavirus-associated Trichodysplasia spinulosa involves hyperproliferation, pRB phosphorylation and upregulation of p16 and p21". PLOS ONE. 9 (10): e108947. Bibcode:2014PLoSO...9j8947K. doi:10.1371/journal.pone.0108947. PMC 4188587. PMID 25291363.
- Kelley WL, Georgopoulos C (April 1997). "The T/t common exon of simian virus 40, JC, and BK polyomavirus T antigens can functionally replace the J-domain of the Escherichia coli DnaJ molecular chaperone". Proceedings of the National Academy of Sciences of the United States of America. 94 (8): 3679–84. Bibcode:1997PNAS...94.3679K. doi:10.1073/pnas.94.8.3679. PMC 20500. PMID 9108037.
- Pallas DC, Shahrik LK, Martin BL, Jaspers S, Miller TB, Brautigan DL, Roberts TM (January 1990). "Polyoma small and middle T antigens and SV40 small t antigen form stable complexes with protein phosphatase 2A". Cell. 60 (1): 167–76. doi:10.1016/0092-8674(90)90726-u. PMID 2153055. S2CID 2007706.
- Sontag E, Fedorov S, Kamibayashi C, Robbins D, Cobb M, Mumby M (December 1993). "The interaction of SV40 small tumor antigen with protein phosphatase 2A stimulates the map kinase pathway and induces cell proliferation". Cell. 75 (5): 887–97. doi:10.1016/0092-8674(93)90533-V. PMID 8252625.
- Watanabe G, Howe A, Lee RJ, Albanese C, Shu IW, Karnezis AN, Zon L, Kyriakis J, Rundell K, Pestell RG (November 1996). "Induction of cyclin D1 by simian virus 40 small tumor antigen". Proceedings of the National Academy of Sciences of the United States of America. 93 (23): 12861–6. Bibcode:1996PNAS...9312861W. doi:10.1073/pnas.93.23.12861. PMC 24011. PMID 8917510.
- Kwun HJ, Shuda M, Feng H, Camacho CJ, Moore PS, Chang Y (August 2013). "Merkel cell polyomavirus small T antigen controls viral replication and oncoprotein expression by targeting the cellular ubiquitin ligase SCFFbw7". Cell Host & Microbe. 14 (2): 125–35. doi:10.1016/j.chom.2013.06.008. PMC 3764649. PMID 23954152.
- Shuda M, Kwun HJ, Feng H, Chang Y, Moore PS (September 2011). "Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator". The Journal of Clinical Investigation. 121 (9): 3623–34. doi:10.1172/JCI46323. PMC 3163959. PMID 21841310.
- Chen XS, Stehle T, Harrison SC (June 1998). "Interaction of polyomavirus internal protein VP2 with the major capsid protein VP1 and implications for participation of VP2 in viral entry". The EMBO Journal. 17 (12): 3233–40. doi:10.1093/emboj/17.12.3233. PMC 1170661. PMID 9628860.
- ^ Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB (June 2010). "Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin". Cell Host & Microbe. 7 (6): 509–15. doi:10.1016/j.chom.2010.05.006. PMC 2919322. PMID 20542254.
- Sariyer IK, Saribas AS, White MK, Safak M (May 2011). "Infection by agnoprotein-negative mutants of polyomavirus JC and SV40 results in the release of virions that are mostly deficient in DNA content". Virology Journal. 8: 255. doi:10.1186/1743-422X-8-255. PMC 3127838. PMID 21609431.
- Saribas AS, Coric P, Hamazaspyan A, Davis W, Axman R, White MK, Abou-Gharbia M, Childers W, Condra JH, Bouaziz S, Safak M (October 2016). "Emerging From the Unknown: Structural and Functional Features of Agnoprotein of Polyomaviruses". Journal of Cellular Physiology. 231 (10): 2115–27. doi:10.1002/jcp.25329. PMC 5217748. PMID 26831433.
- "ICTV Taxonomy Website".
- International Agency for Research on Cancer (2013). "IARC Working Group on the Evaluation of Carcinogenic Risk to Humans. Malaria and Some Polyomaviruses (SV40, BK, JC, and Merkel Cell Viruses)". IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. 104.
- Pérez-Losada M, Christensen RG, McClellan DA, Adams BJ, Viscidi RP, Demma JC, Crandall KA (June 2006). "Comparing phylogenetic codivergence between polyomaviruses and their hosts". Journal of Virology. 80 (12): 5663–9. doi:10.1128/JVI.00056-06. PMC 1472594. PMID 16731904.
- Johne R, Buck CB, Allander T, Atwood WJ, Garcea RL, Imperiale MJ, Major EO, Ramqvist T, Norkin LC (September 2011). "Taxonomical developments in the family Polyomaviridae". Archives of Virology. 156 (9): 1627–34. doi:10.1007/s00705-011-1008-x. PMC 3815707. PMID 21562881.
- ^ "Virus Taxonomy: 2020 Release". International Committee on Taxonomy of Viruses (ICTV). March 2021. Retrieved 10 May 2021.
- Siqueira JD, Ng TF, Miller M, Li L, Deng X, Dodd E, Batac F, Delwart E (July 2017). "Endemic infection of stranded southern sea otters (Enhydra lutris nereis) with novel parvovirus, polyomavirus, and adenovirus". Journal of Wildlife Diseases. 53 (3): 532–542. doi:10.7589/2016-04-082. PMID 28192039. S2CID 46740584.
- Dela Cruz FN, Li L, Delwart E, Pesavento PA (2017). "A novel pulmonary polyomavirus in alpacas (Vicugna pacos)". Veterinary Microbiology. 201: 49–55. doi:10.1016/j.vetmic.2017.01.005. PMID 28284622.
- Qi D, Shan T, Liu Z, Deng X, Zhang Z, Bi W, Owens JR, Feng F, Zheng L, Huang F, Delwart E, Hou R, Zhang W (2017). "A novel polyomavirus from the nasal cavity of a giant panda (Ailuropoda melanoleuca)". Virology Journal. 14 (1): 207. doi:10.1186/s12985-017-0867-5. PMC 5658932. PMID 29078783.
- Gonçalves Motta Maia F, Marciel de Souza W, Sabino-Santos G, Jorge Fumagalli M, Modha S, Ramiro Murcia P, Tadeu Moraes Figueiredo L (2018). "A novel polyomavirus in sigmodontine rodents from São Paulo State, Brazil". Archives of Virology. 163 (10): 2913–2915. doi:10.1007/s00705-018-3913-8. PMID 29931397. S2CID 49351836.
- Liu P, Qiu Y, Xing C, Zhou JH, Yang WH, Wang Q, Li JY, Han X, Zhang YZ, Ge XY (2019). "Detection and genome characterization of two novel papillomaviruses and a novel polyomavirus in tree shrew (Tupaia belangeri chinensis) in China". Virol J. 16 (1): 35. doi:10.1186/s12985-019-1141-9. PMC 6423848. PMID 30885224.
- Gheit T, Dutta S, Oliver J, Robitaille A, Hampras S, Combes JD, McKay-Chopin S, Le Calvez-Kelm F, Fenske N, Cherpelis B, Giuliano AR, Franceschi S, McKay J, Rollison DE, Tommasino M (2017). "Isolation and characterization of a novel putative human polyomavirus". Virology. 506: 45–54. doi:10.1016/j.virol.2017.03.007. PMC 9265179. PMID 28342387.
- Altman LK (18 January 2008). "Virus Is Linked to a Powerful Skin Cancer". The New York Times. Retrieved 18 January 2008.
- Shuda M, Arora R, Kwun HJ, Feng H, Sarid R, Fernández-Figueras MT, Tolstov Y, Gjoerup O, Mansukhani MM, Swerdlow SH, Chaudhary PM, Kirkwood JM, Nalesnik MA, Kant JA, Weiss LM, Moore PS, Chang Y (September 2009). "Human Merkel cell polyomavirus infection I. MCV T antigen expression in Merkel cell carcinoma, lymphoid tissues and lymphoid tumors". International Journal of Cancer. 125 (6): 1243–9. doi:10.1002/ijc.24510. PMC 6388400. PMID 19499546.
- van der Meijden E, Janssens RW, Lauber C, Bouwes Bavinck JN, Gorbalenya AE, Feltkamp MC (July 2010). "Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient". PLOS Pathogens. 6 (7): e1001024. doi:10.1371/journal.ppat.1001024. PMC 2912394. PMID 20686659.
- Kazem S, van der Meijden E, Feltkamp MC (August 2013). "The trichodysplasia spinulosa-associated polyomavirus: virological background and clinical implications". APMIS. 121 (8): 770–82. doi:10.1111/apm.12092. PMID 23593936. S2CID 13734654.
- Scuda N, Hofmann J, Calvignac-Spencer S, Ruprecht K, Liman P, Kühn J, Hengel H, Ehlers B (May 2011). "A novel human polyomavirus closely related to the african green monkey-derived lymphotropic polyomavirus". Journal of Virology. 85 (9): 4586–90. doi:10.1128/jvi.02602-10. PMC 3126223. PMID 21307194.
- Korup S, Rietscher J, Calvignac-Spencer S, Trusch F, Hofmann J, Moens U, Sauer I, Voigt S, Schmuck R, Ehlers B (2013). "Identification of a novel human polyomavirus in organs of the gastrointestinal tract". PLOS ONE. 8 (3): e58021. Bibcode:2013PLoSO...858021K. doi:10.1371/journal.pone.0058021. PMC 3596337. PMID 23516426.
- Mishra N, Pereira M, Rhodes RH, An P, Pipas JM, Jain K, Kapoor A, Briese T, Faust PL, Lipkin WI (November 2014). "Identification of a novel polyomavirus in a pancreatic transplant recipient with retinal blindness and vasculitic myopathy". The Journal of Infectious Diseases. 210 (10): 1595–9. doi:10.1093/infdis/jiu250. PMC 4334791. PMID 24795478.
- Gardner SD, Field AM, Coleman DV, Hulme B (June 1971). "New human papovavirus (B.K.) isolated from urine after renal transplantation". Lancet. 1 (7712): 1253–7. doi:10.1016/s0140-6736(71)91776-4. PMID 4104714.
- Padgett BL, Walker DL, ZuRhein GM, Eckroade RJ, Dessel BH (June 1971). "Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy". Lancet. 1 (7712): 1257–60. doi:10.1016/S0140-6736(71)91777-6. PMID 4104715.
- Allander T, Andreasson K, Gupta S, Bjerkner A, Bogdanovic G, Persson MA, Dalianis T, Ramqvist T, Andersson B (April 2007). "Identification of a third human polyomavirus". Journal of Virology. 81 (8): 4130–6. doi:10.1128/JVI.00028-07. PMC 1866148. PMID 17287263.
- ^ Nguyen KD, Lee EE, Yue Y, Stork J, Pock L, North JP, Vandergriff T, Cockerell C, Hosler GA, Pastrana DV, Buck CB, Wang RC (May 2017). "Human polyomavirus 6 and 7 are associated with pruritic and dyskeratotic dermatoses". Journal of the American Academy of Dermatology. 76 (5): 932–940.e3. doi:10.1016/j.jaad.2016.11.035. PMC 5392424. PMID 28040372.
- Ho J, Jedrych JJ, Feng H, Natalie AA, Grandinetti L, Mirvish E, Crespo MM, Yadav D, Fasanella KE, Proksell S, Kuan SF, Pastrana DV, Buck CB, Shuda Y, Moore PS, Chang Y (May 2015). "Human polyomavirus 7-associated pruritic rash and viremia in transplant recipients". The Journal of Infectious Diseases. 211 (10): 1560–5. doi:10.1093/infdis/jiu524. PMC 4425822. PMID 25231015.
- Toptan T, Yousem SA, Ho J, Matsushima Y, Stabile LP, Fernández-Figueras MT, Bhargava R, Ryo A, Moore PS, Chang Y (February 2016). "Survey for human polyomaviruses in cancer". JCI Insight. 1 (2). doi:10.1172/jci.insight.85562. PMC 4811373. PMID 27034991.
- Siebrasse EA, Reyes A, Lim ES, Zhao G, Mkakosya RS, Manary MJ, Gordon JI, Wang D (October 2012). "Identification of MW polyomavirus, a novel polyomavirus in human stool". Journal of Virology. 86 (19): 10321–6. doi:10.1128/JVI.01210-12. PMC 3457274. PMID 22740408.
- Buck CB, Phan GQ, Raiji MT, Murphy PM, McDermott DH, McBride AA (October 2012). "Complete genome sequence of a tenth human polyomavirus". Journal of Virology. 86 (19): 10887. doi:10.1128/JVI.01690-12. PMC 3457262. PMID 22966183.
- Yu G, Greninger AL, Isa P, Phan TG, Martínez MA, de la Luz Sanchez M, Contreras JF, Santos-Preciado JI, Parsonnet J, Miller S, DeRisi JL, Delwart E, Arias CF, Chiu CY (2012). "Discovery of a novel polyomavirus in acute diarrheal samples from children". PLOS ONE. 7 (11): e49449. Bibcode:2012PLoSO...749449Y. doi:10.1371/journal.pone.0049449. PMC 3498111. PMID 23166671.
- Lim ES, Reyes A, Antonio M, Saha D, Ikumapayi UN, Adeyemi M, Stine OC, Skelton R, Brennan DC, Mkakosya RS, Manary MJ, Gordon JI, Wang D (February 2013). "Discovery of STL polyomavirus, a polyomavirus of ancestral recombinant origin that encodes a unique T antigen by alternative splicing". Virology. 436 (2): 295–303. doi:10.1016/j.virol.2012.12.005. PMC 3693558. PMID 23276405.
- Gheit, Tarik; Dutta, Sankhadeep; Oliver, Javier; Robitaille, Alexis; Hampras, Shalaka; Combes, Jean-Damien; McKay-Chopin, Sandrine; Calvez-Kelm, Florence Le; Fenske, Neil (2017). "Isolation and characterization of a novel putative human polyomavirus". Virology. 506: 45–54. doi:10.1016/j.virol.2017.03.007. PMC 9265179. PMID 28342387.
- Prado JC, Monezi TA, Amorim AT, Lino V, Paladino A, Boccardo E (2018). "Human polyomaviruses and cancer: an overview". Clinics (Sao Paulo). 73 (suppl 1): e558s. doi:10.6061/clinics/2018/e558s. PMC 6157077. PMID 30328951.
- Dalianis T, Hirsch HH (March 2013). "Human polyomaviruses in disease and cancer". Virology. 437 (2): 63–72. doi:10.1016/j.virol.2012.12.015. PMID 23357733.
- Van Ghelue M, Khan MT, Ehlers B, Moens U (November 2012). "Genome analysis of the new human polyomaviruses". Reviews in Medical Virology. 22 (6): 354–77. doi:10.1002/rmv.1711. PMID 22461085.
- Egli A, Infanti L, Dumoulin A, Buser A, Samaridis J, Stebler C, Gosert R, Hirsch HH (March 2009). "Prevalence of polyomavirus BK and JC infection and replication in 400 healthy blood donors". The Journal of Infectious Diseases. 199 (6): 837–46. doi:10.1086/597126. PMID 19434930.
- ^ Poulin DL, DeCaprio JA (September 2006). "Is there a role for SV40 in human cancer?". Journal of Clinical Oncology. 24 (26): 4356–65. doi:10.1200/JCO.2005.03.7101. PMID 16963733.
- ^ zur Hausen H (December 2003). "SV40 in human cancers--an endless tale?". International Journal of Cancer. 107 (5): 687. doi:10.1002/ijc.11517. PMID 14566815.
- Gazdar AF, Butel JS, Carbone M (December 2002). "SV40 and human tumours: myth, association or causality?". Nature Reviews. Cancer. 2 (12): 957–64. doi:10.1038/nrc947. PMID 12459734. S2CID 8878662.
- Carroll-Pankhurst C, Engels EA, Strickler HD, Goedert JJ, Wagner J, Mortimer EA (November 2001). "Thirty-five year mortality following receipt of SV40- contaminated polio vaccine during the neonatal period". British Journal of Cancer. 85 (9): 1295–7. doi:10.1054/bjoc.2001.2065. PMC 2375249. PMID 11720463.
- Shah KV (January 2007). "SV40 and human cancer: a review of recent data". International Journal of Cancer. 120 (2): 215–23. doi:10.1002/ijc.22425. PMID 17131333. S2CID 20679358.
- ^ Drachenberg CB, Hirsch HH, Ramos E, Papadimitriou JC (December 2005). "Polyomavirus disease in renal transplantation: review of pathological findings and diagnostic methods". Human Pathology. 36 (12): 1245–55. doi:10.1016/j.humpath.2005.08.009. PMID 16311117.
- Viscidi RP, Clayman B (2006). "Serological Cross Reactivity between Polyomavirus Capsids". In Ahsan N (ed.). Polyomaviruses and Human Diseases. Advances in Experimental Medicine and Biology. Vol. 577. pp. 73–84. doi:10.1007/0-387-32957-9_5. ISBN 978-0-387-29233-5. PMID 16626028.
- Drews K, Bashir T, Dörries K (January 2000). "Quantification of human polyomavirus JC in brain tissue and cerebrospinal fluid of patients with progressive multifocal leukoencephalopathy by competitive PCR". Journal of Virological Methods. 84 (1): 23–36. doi:10.1016/S0166-0934(99)00128-7. PMID 10644084.
- Nickeleit V, Hirsch HH, Binet IF, Gudat F, Prince O, Dalquen P, Thiel G, Mihatsch MJ (May 1999). "Polyomavirus infection of renal allograft recipients: from latent infection to manifest disease". Journal of the American Society of Nephrology. 10 (5): 1080–9. doi:10.1681/ASN.V1051080. PMID 10232695.
- Randhawa PS, Vats A, Zygmunt D, Swalsky P, Scantlebury V, Shapiro R, Finkelstein S (August 2002). "Quantitation of viral DNA in renal allograft tissue from patients with BK virus nephropathy". Transplantation. 74 (4): 485–8. doi:10.1097/00007890-200208270-00009. PMID 12352906. S2CID 30574884.
- Busam KJ, Jungbluth AA, Rekthman N, Coit D, Pulitzer M, Bini J, Arora R, Hanson NC, Tassello JA, Frosina D, Moore P, Chang Y (September 2009). "Merkel cell polyomavirus expression in merkel cell carcinomas and its absence in combined tumors and pulmonary neuroendocrine carcinomas". The American Journal of Surgical Pathology. 33 (9): 1378–85. doi:10.1097/PAS.0b013e3181aa30a5. PMC 2932664. PMID 19609205.
- Tolstov YL, Pastrana DV, Feng H, Becker JC, Jenkins FJ, Moschos S, Chang Y, Buck CB, Moore PS (September 2009). "Human Merkel cell polyomavirus infection II. MCV is a common human infection that can be detected by conformational capsid epitope immunoassays". International Journal of Cancer. 125 (6): 1250–6. doi:10.1002/ijc.24509. PMC 2747737. PMID 19499548.
- Pastrana DV, Tolstov YL, Becker JC, Moore PS, Chang Y, Buck CB (September 2009). "Quantitation of human seroresponsiveness to Merkel cell polyomavirus". PLOS Pathogens. 5 (9): e1000578. doi:10.1371/journal.ppat.1000578. PMC 2734180. PMID 19750217.
- Carter JJ, Paulson KG, Wipf GC, Miranda D, Madeleine MM, Johnson LG, Lemos BD, Lee S, Warcola AH, Iyer JG, Nghiem P, Galloway DA (November 2009). "Association of Merkel cell polyomavirus-specific antibodies with Merkel cell carcinoma". Journal of the National Cancer Institute. 101 (21): 1510–22. doi:10.1093/jnci/djp332. PMC 2773184. PMID 19776382.
- Elizabeth Matisoo-Smith; K. Ann Horsburgh (2012). DNA for Archaeologists. Routledge. ISBN 978-1598746815.
- Sugimoto C, Kitamura T, Guo J, Al-Ahdal MN, Shchelkunov SN, Otova B, Ondrejka P, Chollet JY, El-Safi S, Ettayebi M, Grésenguet G, Kocagöz T, Chaiyarasamee S, Thant KZ, Thein S, Moe K, Kobayashi N, Taguchi F, Yogo Y (19 August 1997). "Typing of urinary JC virus DNA offers a novel means of tracing human migrations". Proc Natl Acad Sci U S A. 94 (17): 9191–9196. Bibcode:1997PNAS...94.9191S. doi:10.1073/pnas.94.17.9191. PMC 23108. PMID 9256458.
- Gross L (June 1953). "A filterable agent, recovered from Ak leukemic extracts, causing salivary gland carcinomas in C3H mice". Proceedings of the Society for Experimental Biology and Medicine. 83 (2): 414–21. doi:10.3181/00379727-83-20376. PMID 13064287. S2CID 34223353.
- Stewart SE, Eddy BE, Borgese N (June 1958). "Neoplasms in mice inoculated with a tumor agent carried in tissue culture". Journal of the National Cancer Institute. 20 (6): 1223–43. doi:10.1093/jnci/20.6.1223. PMID 13549981.
- Eddy BE, Stewart SE (November 1959). "Characteristics of the SE polyoma virus". American Journal of Public Health and the Nation's Health. 49 (11): 1486–92. doi:10.2105/AJPH.49.11.1486. PMC 1373056. PMID 13819251.
- Schowalter RM, Pastrana DV, Buck CB (July 2011). "Glycosaminoglycans and sialylated glycans sequentially facilitate Merkel cell polyomavirus infectious entry". PLOS Pathogens. 7 (7): e1002161. doi:10.1371/journal.ppat.1002161. PMC 3145800. PMID 21829355.
- Gottlieb KA, Villarreal LP (June 2001). "Natural biology of polyomavirus middle T antigen". Microbiology and Molecular Biology Reviews. 65 (2): 288–318, second and third pages, table of contents. doi:10.1128/mmbr.65.2.288-318.2001. PMC 99028. PMID 11381103.
- Peretti A, FitzGerald PC, Bliskovsky V, Pastrana DV, Buck CB (January 2015). "Genome Sequence of a Fish-Associated Polyomavirus, Black Sea Bass (Centropristis striata) Polyomavirus 1". Genome Announcements. 3 (1): e01476-14. doi:10.1128/genomeA.01476-14. PMC 4319505. PMID 25635011.
- López-Bueno A, Mavian C, Labella AM, Castro D, Borrego JJ, Alcami A, Alejo A (October 2016). "Concurrence of Iridovirus, Polyomavirus, and a Unique Member of a New Group of Fish Papillomaviruses in Lymphocystis Disease-Affected Gilthead Sea Bream". Journal of Virology. 90 (19): 8768–79. doi:10.1128/JVI.01369-16. PMC 5021401. PMID 27440877.
External links
| Taxon identifiers | |
|---|---|
| Polyomaviridae | |