| Revision as of 13:39, 13 May 2020 editCitation bot (talk | contribs)Bots5,445,338 edits Alter: template type. Add: isbn, pmc, pmid, pages, volume, year, series, title, chapter, author pars. 1-2. Formatted dashes. | You can use this bot yourself. Report bugs here. | Activated by Chris Capoccia | via #UCB_toolbar← Previous edit | Latest revision as of 14:29, 29 October 2024 edit undo2601:781:4200:de90:10d7:106c:a31f:d486 (talk)No edit summary | ||
| (35 intermediate revisions by 23 users not shown) | |||
| Line 1: | Line 1: | ||
| {{short description|Class of chemical compounds}} | {{short description|Class of chemical compounds}} | ||
| ⚫ | A '''nucleoside triphosphate''' is a |
||
| ⚫ | A '''nucleoside triphosphate''' is a ] containing a ] bound to a 5-carbon sugar (either ] or ]), with three ]s bound to the sugar.<ref name=":0">{{Cite news|url=http://knowgenetics.org/nucleotides-and-bases/|title=Nucleotides and Bases - Genetics Generation|work=Genetics Generation | access-date = 11 November 2017 }}</ref> They are the molecular precursors of both ] and ], which are chains of ]s made through the processes of ] and ].<ref name=":8">{{cite book | url = https://books.google.com/books?id=o0xA2udqufwC&q=nucleic+acid+synthesis&pg=PP1 | title = The Nucleic Acids | vauthors = Chargaff E | date=2012-12-02|publisher=Elsevier|isbn=9780323144773}}</ref> Nucleoside triphosphates also serve as a source of energy for cellular reactions<ref name=Khan-ATP>{{Cite web|url=https://www.khanacademy.org/test-prep/mcat/biomolecules/overview-metabolism/a/overview-of-atp-hydrolysis|title=Overview of ATP Hydrolysis|website=Khan Academy|archive-url=https://web.archive.org/web/20171201042725/https://www.khanacademy.org/test-prep/mcat/biomolecules/overview-metabolism/a/overview-of-atp-hydrolysis|archive-date=2017-12-01|access-date=2017-11-11|url-status=dead}}</ref> and are involved in signalling pathways.<ref name=gpcr14047471>{{cite web |title=GPCR |work=Scitable |date=2014 |url=https://www.nature.com/scitable/topicpage/gpcr-14047471/ }}</ref> | ||
| ⚫ | Nucleoside triphosphates cannot |
||
| ⚫ | Nucleoside triphosphates cannot easily cross the cell membrane, so they are typically synthesized within the cell.<ref>{{Cite news|url=https://honey-guide.com/2014/04/09/eating-dna-dietary-nucleotides-in-nutrition/|title=Eating DNA: Dietary Nucleotides in Nutrition|date=2014-04-09|work=The call of the Honeyguide|access-date=11 November 2017}}</ref> Synthesis pathways differ depending on the specific nucleoside triphosphate being made, but given the many important roles of nucleoside triphosphates, synthesis is tightly regulated in all cases.<ref name="Wyngaarden_1976">{{cite journal | vauthors = Wyngaarden JB | title = Regulation of purine biosynthesis and turnover | journal = Advances in Enzyme Regulation | volume = 14 | pages = 25–42 | year = 1976 | pmid = 184697 | doi = 10.1016/0065-2571(76)90006-6 }}</ref> ]s may also be used to treat viral infections.<ref name="Galmarini_2001"/> For example, ] (AZT) is a nucleoside analogue used to prevent and treat ].<ref name=":10">{{Cite news|url=https://www.drugs.com/monograph/zidovudine.html|title=Zidovudine Monograph for Professionals - Drugs.com|work=Drugs.com|access-date= 30 November 2017 }}</ref> | ||
| == Naming == | == Naming == | ||
| The term ] refers to a ] linked to a 5-carbon sugar (either ] or ]).<ref name=":0" /> ]s are nucleosides ] linked to one or more ]s.<ref>{{cite book| |
The term ] refers to a ] linked to a 5-carbon sugar (either ] or ]).<ref name=":0" /> ]s are nucleosides ] linked to one or more ]s.<ref>{{cite book| vauthors = Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J |date=2000|title=Structure of Nucleic Acids|url=https://www.ncbi.nlm.nih.gov/books/NBK21514/}}</ref> To provide information about the number of phosphates, nucleotides may instead be referred to as nucleoside (mono, di, or tri) phosphates.<ref name=":1">{{cite journal | vauthors = Secrist JA | title = Nucleoside and nucleotide nomenclature | journal = Current Protocols in Nucleic Acid Chemistry | volume = Appendix 1 | pages = A.1D.1–A.1D.3 | date = May 2001 | pmid = 18428808 | doi = 10.1002/0471142700.nca01ds00 | hdl = 2027.42/143595 | s2cid = 205152902 | url = https://deepblue.lib.umich.edu/bitstream/2027.42/143595/1/cpnca01d.pdf | hdl-access = free }}</ref> Thus, nucleoside triphosphates are a type of nucleotide.<ref name=":1" /> | ||
| Nucleotides are commonly abbreviated with 3 letters (4 or 5 in case of deoxy- or dideoxy-nucleotides). The first letter indicates the identity of the nitrogenous base (e.g. A for ], G for ]), the second letter indicates the number of phosphates (mono, di, tri), and the third letter is P, standing for phosphate.<ref>{{Cite web|url=http://www.biochem.uthscsa.edu/med/02-Nucleotide-Metab/PrereqNucleotideMetab3.html|title=Nomenclature of Nucleosides|website=www.biochem.uthscsa.edu|access-date=2017-11-11}}</ref> Nucleoside triphosphates that contain ] as the sugar are conventionally abbreviated as NTPs, while nucleoside triphosphates containing ] as the sugar are abbreviated as dNTPs. For example, dATP stands for deoxyribose |
Nucleotides are commonly abbreviated with 3 letters (4 or 5 in case of deoxy- or dideoxy-nucleotides). The first letter indicates the identity of the nitrogenous base (e.g., A for ], G for ]), the second letter indicates the number of phosphates (mono, di, tri), and the third letter is P, standing for phosphate.<ref>{{Cite web|url=http://www.biochem.uthscsa.edu/med/02-Nucleotide-Metab/PrereqNucleotideMetab3.html|title=Nomenclature of Nucleosides|website=www.biochem.uthscsa.edu|access-date=2017-11-11}}</ref> Nucleoside triphosphates that contain ] as the sugar are conventionally abbreviated as NTPs, while nucleoside triphosphates containing ] as the sugar are abbreviated as dNTPs. For example, dATP stands for deoxyribose adenosine triphosphate. NTPs are the building blocks of ], and dNTPs are the building blocks of ].<ref>{{Cite news|url=https://science-explained.com/theory/dna-rna-and-protein/|title=From DNA to RNA to protein, how does it work?|work=Science Explained|access-date=11 November 2017}}</ref> | ||
| The carbons of the sugar in a nucleoside triphosphate are numbered around the carbon ring starting from the original ] of the sugar. Conventionally, the carbon numbers in a sugar are followed by the prime symbol (‘) to distinguish them from the carbons of the nitrogenous base. The nitrogenous base is linked to the 1’ carbon through a ], and the phosphate groups are covalently linked to the 5’ carbon.<ref>{{Cite web|url=http://www.biosyn.com/tew/numbering-convention-for-nucleotides.aspx|title=Numbering convention for nucleotides |
The carbons of the sugar in a nucleoside triphosphate are numbered around the carbon ring starting from the original ] of the sugar. Conventionally, the carbon numbers in a sugar are followed by the prime symbol (‘) to distinguish them from the carbons of the nitrogenous base. The nitrogenous base is linked to the 1’ carbon through a ], and the phosphate groups are covalently linked to the 5’ carbon.<ref>{{Cite web|url=http://www.biosyn.com/tew/numbering-convention-for-nucleotides.aspx|title=Numbering convention for nucleotides |website=www.biosyn.com|access-date=2017-11-11}}</ref> The first phosphate group linked to the sugar is termed the α-phosphate, the second is the β-phosphate, and the third is the γ-phosphate; these are linked to one another by two ] bonds.<ref>{{cite web | url = http://www.sparknotes.com/biology/molecular/dnareplicationandrepair/section2.rhtml|title=SparkNotes: DNA Replication and Repair: The Chemistry of the Addition of Substrates of DNA Replication|website=www.sparknotes.com|access-date=2017-11-11}}</ref>] | ||
| == DNA and RNA synthesis == | == DNA and RNA synthesis == | ||
| ]The cellular processes of ] and ] involve DNA and RNA synthesis, respectively. DNA synthesis uses dNTPs as substrates, while RNA synthesis uses |
]The cellular processes of ] and ] involve DNA and RNA synthesis, respectively. DNA synthesis uses dNTPs as substrates, while RNA synthesis uses rNTPs as substrates.<ref name=":8" /> NTPs cannot be converted directly to dNTPs. DNA contains four different nitrogenous bases: ], ], ] and ]. RNA also contains adenine, guanine, and cytosine, but replaces thymine with ].<ref>{{Cite news|url=https://www.thoughtco.com/dna-versus-rna-608191|title=Do You Know the Differences Between DNA and RNA?|work=ThoughtCo|access-date=2017-11-11}}</ref> Thus, DNA synthesis requires dATP, dGTP, dCTP, and dTTP as substrates, while RNA synthesis requires ATP, GTP, CTP, and UTP. | ||
| Nucleic acid synthesis is catalyzed by either ] or ] for DNA and RNA synthesis respectively.<ref>{{Cite web|url=http://www.differencebetween.com/difference-between-dna-polymerase-and-vs-rna-polymerase/|title=Difference Between DNA Polymerase and RNA Polymerase |
Nucleic acid synthesis is catalyzed by either ] or ] for DNA and RNA synthesis respectively.<ref>{{Cite web|url=http://www.differencebetween.com/difference-between-dna-polymerase-and-vs-rna-polymerase/|title=Difference Between DNA Polymerase and RNA Polymerase|access-date=2017-11-11|date=2011-12-24}}</ref> These enzymes ] link the free ] group on the 3’ carbon of a growing chain of nucleotides to the α-phosphate on the 5’ carbon of the next (d)NTP, releasing the β- and γ-phosphate groups as ] (PP<sub>i</sub>).<ref name=":2">{{cite book | vauthors = Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J |date=2000|title=Nucleic Acid Synthesis|url=https://www.ncbi.nlm.nih.gov/books/NBK21644/}}</ref> This results in a ] linkage between the two (d)NTPs. The release of PP<sub>i</sub> provides the energy necessary for the reaction to occur.<ref name=":2" /> It is important to note that nucleic acid synthesis occurs exclusively in the ]. | ||
| == Nucleoside triphosphate metabolism == | == Nucleoside triphosphate metabolism == | ||
| Given their importance in the cell, the synthesis and degradation of nucleoside triphosphates is under tight control.<ref name="Wyngaarden_1976" /> This section focuses on nucleoside triphosphate metabolism in humans, but the process is fairly conserved among species.<ref>{{cite journal | vauthors = Samant S, Lee H, Ghassemi M, Chen J, Cook JL, Mankin AS, Neyfakh AA | title = Nucleotide biosynthesis is critical for growth of bacteria in human blood | journal = PLOS Pathogens | volume = 4 | issue = 2 | pages = e37 | date = February 2008 | pmid = 18282099 | doi = 10.1371/journal.ppat.0040037 | pmc=2242838}}</ref> Nucleoside triphosphates cannot be absorbed well, so all nucleoside triphosphates are typically made '']''.<ref>{{cite book | |
Given their importance in the cell, the synthesis and degradation of nucleoside triphosphates is under tight control.<ref name="Wyngaarden_1976" /> This section focuses on nucleoside triphosphate metabolism in humans, but the process is fairly conserved among species.<ref>{{cite journal | vauthors = Samant S, Lee H, Ghassemi M, Chen J, Cook JL, Mankin AS, Neyfakh AA | title = Nucleotide biosynthesis is critical for growth of bacteria in human blood | journal = PLOS Pathogens | volume = 4 | issue = 2 | pages = e37 | date = February 2008 | pmid = 18282099 | doi = 10.1371/journal.ppat.0040037 | pmc=2242838 | doi-access = free }}</ref> Nucleoside triphosphates cannot be absorbed well, so all nucleoside triphosphates are typically made '']''.<ref>{{cite book | vauthors = Berg JM, Tymoczko JL, Stryer L |date=2002|title=Nucleotide Biosynthesis|url=https://www.ncbi.nlm.nih.gov/books/NBK21216/}}</ref> The synthesis of ATP and GTP (]s) differs from the synthesis of CTP, TTP, and UTP (]s). Both purine and pyrimidine synthesis use ] (PRPP) as a starting molecule.<ref name=":5">{{cite web | url = https://themedicalbiochemistrypage.org/nucleotide-metabolism.php|title=Nucleotide Metabolism: Nucleic Acid Synthesis|website=themedicalbiochemistrypage.org|access-date=2017-11-15}}</ref> | ||
| The conversion of NTPs to dNTPs can only be done in the diphosphate form. Typically a NTP has one phosphate removed to become a NDP, then is converted to a dNDP by an enzyme called ], then a phosphate is added back to give a dNTP.<ref name="Stubbe_1990">{{cite journal | vauthors = Stubbe J | title = Ribonucleotide reductases: amazing and confusing | journal = The Journal of Biological Chemistry | volume = 265 | issue = 10 | pages = 5329–32 | year = 1990 | |
The conversion of NTPs to dNTPs can only be done in the diphosphate form. Typically a NTP has one phosphate removed to become a NDP, then is converted to a dNDP by an enzyme called ], then a phosphate is added back to give a dNTP.<ref name="Stubbe_1990">{{cite journal | vauthors = Stubbe J | title = Ribonucleotide reductases: amazing and confusing | journal = The Journal of Biological Chemistry | volume = 265 | issue = 10 | pages = 5329–32 | year = 1990 | doi = 10.1016/S0021-9258(19)39357-3 | pmid = 2180924 | url = http://www.jbc.org/content/265/10/5329.full.pdf | doi-access = free }}</ref> | ||
| === Purine synthesis === | === Purine synthesis === | ||
| A nitrogenous base called ] is assembled directly onto PRPP.<ref>{{cite book| |
A nitrogenous base called ] is assembled directly onto PRPP.<ref>{{cite book| vauthors = Berg J, Tymoczko JL, Stryer L |date=2002 |title=Purine Bases Can Be Synthesized de Novo or Recycled by Salvage Pathways|url=https://www.ncbi.nlm.nih.gov/books/NBK22385/}}</ref> This results in a nucleotide called ] (IMP). IMP is then converted to either a precursor to AMP or GMP. Once AMP or GMP are formed, they can be phosphorylated by ATP to their diphosphate and triphosphate forms.<ref>{{Cite news|url=http://www.biochemden.com/purine-synthesis/|title=Purine Synthesis : Synthesis of Purine RiboNucleotides|date=2016-03-16|work=BiochemDen.com|access-date= 15 November 2017 }}</ref> | ||
| Purine synthesis is regulated by the ] of IMP formation by the adenine or guanine nucleotides.<ref>{{cite book| |
Purine synthesis is regulated by the ] of IMP formation by the adenine or guanine nucleotides.<ref>{{cite book | vauthors = Berg JM, Tymoczko JL, Stryer L |date=2002|title=Key Steps in Nucleotide Biosynthesis Are Regulated by Feedback Inhibition|url=https://www.ncbi.nlm.nih.gov/books/NBK22428/}}</ref> AMP and GMP also ] the formation of their precursors from IMP.<ref name="Nierlich_1965">{{cite journal | vauthors = Nierlich DP, Magasanik B | title = Regulation of purine ribonucleotide synthesis by end product inhibition. the effect of adenine and guanine ribonucleotides on the 5'-phosphoribosyl-pyrophosphate amidotransferase of aerobacter aerogenes | journal = The Journal of Biological Chemistry | volume = 240 | pages = 358–65 | year = 1965 | doi = 10.1016/S0021-9258(18)97657-X | pmid = 14253438 | doi-access = free }}</ref> | ||
| === Pyrimidine synthesis === | === Pyrimidine synthesis === | ||
| A nitrogenous base called ] is synthesized independently of PRPP.<ref name="Nierlich_1965" /> After orotate is made it is covalently attached to PRPP. This results in a nucleotide called orotate monophosphate (OMP).<ref>{{cite journal | vauthors = Moffatt BA, Ashihara H | title = Purine and pyrimidine nucleotide synthesis and metabolism | journal = The Arabidopsis Book | volume = 1 | pages = e0018 | date = April 2002 | pmid = 22303196 | pmc = 3243375 | doi = 10.1199/tab.0018 }}</ref> OMP is converted to UMP, which can then be phosphorylated by ATP to UDP and UTP. UTP can then be converted to CTP by a ] reaction.<ref>{{cite web|url=https://www.cliffsnotes.com/study-guides/biology/biochemistry-ii/purines-and-pyrimidines/pyrimidine-metabolism|title=Pyrimidine Metabolism|website=www.cliffsnotes.com|access-date=2017-11-15}}</ref> TTP is not a substrate for nucleic acid synthesis, so it is not synthesized in the cell. Instead, dTTP is made indirectly from either dUDP or dCDP after conversion to their deoxyribose forms.<ref name=":5" /> | A nitrogenous base called ] is synthesized independently of PRPP.<ref name="Nierlich_1965" /> After orotate is made it is covalently attached to PRPP. This results in a nucleotide called orotate monophosphate (OMP).<ref>{{cite journal | vauthors = Moffatt BA, Ashihara H | title = Purine and pyrimidine nucleotide synthesis and metabolism | journal = The Arabidopsis Book | volume = 1 | pages = e0018 | date = April 2002 | pmid = 22303196 | pmc = 3243375 | doi = 10.1199/tab.0018 }}</ref> OMP is converted to UMP, which can then be phosphorylated by ATP to UDP and UTP. UTP can then be converted to CTP by a ] reaction.<ref>{{cite web|url=https://www.cliffsnotes.com/study-guides/biology/biochemistry-ii/purines-and-pyrimidines/pyrimidine-metabolism|title=Pyrimidine Metabolism|website=www.cliffsnotes.com|access-date=2017-11-15}}</ref> TTP is not a substrate for nucleic acid synthesis, so it is not synthesized in the cell. Instead, dTTP is made indirectly from either dUDP or dCDP after conversion to their respective deoxyribose forms.<ref name=":5" /> | ||
| Pyrimidine synthesis is regulated by the allosteric inhibition of orotate synthesis by UDP and UTP. PRPP and ATP are also allosteric activators of orotate synthesis.<ref>{{cite journal | vauthors = Lane AN, Fan TW | title = Regulation of mammalian nucleotide metabolism and biosynthesis | journal = Nucleic Acids Research | volume = 43 | issue = 4 | pages = 2466–85 | date = February 2015 | pmid = 25628363 | doi = 10.1093/nar/gkv047 | pmc=4344498}}</ref> | Pyrimidine synthesis is regulated by the allosteric inhibition of orotate synthesis by UDP and UTP. PRPP and ATP are also allosteric activators of orotate synthesis.<ref>{{cite journal | vauthors = Lane AN, Fan TW | title = Regulation of mammalian nucleotide metabolism and biosynthesis | journal = Nucleic Acids Research | volume = 43 | issue = 4 | pages = 2466–85 | date = February 2015 | pmid = 25628363 | doi = 10.1093/nar/gkv047 | pmc=4344498}}</ref> | ||
| === Ribonucleotide reductase === | === Ribonucleotide reductase === | ||
| ] (RNR) is the enzyme responsible for converting NTPs to dNTPs. Given that dNTPs are used in DNA replication, the activity of RNR is tightly regulated.<ref name="Wyngaarden_1976" /> It is important to note that RNR can only process NDPs, so NTPs are first dephosphorylated to NDPs before conversion to dNDPs.<ref name="auto">{{cite journal | vauthors = Kolberg M, Strand KR, Graff P, Andersson KK | title = Structure, function, and mechanism of ribonucleotide reductases | journal = Biochimica et Biophysica Acta | volume = 1699 | issue = 1–2 | pages = 1–34 | date = June 2004 | pmid = 15158709 | doi = 10.1016/j.bbapap.2004.02.007 }}</ref> dNDPs are then typically re-phosphorylated. RNR has 2 subunits and 3 sites: the catalytic site, activity (A) site, and specificity (S) site.<ref name="auto"/> The catalytic site is where the NDP to dNDP reaction takes place, the activity site determines whether or not the enzyme is active, and the specificity site determines which reaction takes place in the catalytic site. | ] (RNR) is the enzyme responsible for converting NTPs to dNTPs. Given that dNTPs are used in DNA replication, the activity of RNR is tightly regulated.<ref name="Wyngaarden_1976" /> It is important to note that RNR can only process NDPs, so NTPs are first dephosphorylated to NDPs before conversion to dNDPs.<ref name="auto">{{cite journal | vauthors = Kolberg M, Strand KR, Graff P, Andersson KK | title = Structure, function, and mechanism of ribonucleotide reductases | journal = Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics | volume = 1699 | issue = 1–2 | pages = 1–34 | date = June 2004 | pmid = 15158709 | doi = 10.1016/j.bbapap.2004.02.007 }}</ref> dNDPs are then typically re-phosphorylated. RNR has 2 subunits and 3 sites: the catalytic site, activity (A) site, and specificity (S) site.<ref name="auto"/> The catalytic site is where the NDP to dNDP reaction takes place, the activity site determines whether or not the enzyme is active, and the specificity site determines which reaction takes place in the catalytic site. | ||
| The activity site can bind either ATP or dATP.<ref name=":7">{{cite book |doi=10.1016/B978-0-12-386931-9.00014-3 |chapter=The Structural Basis for the Allosteric Regulation of Ribonucleotide Reductase |title=Oligomerization in Health and Disease |series=Progress in Molecular Biology and Translational Science |year=2013 | |
The activity site can bind either ATP or dATP.<ref name=":7">{{cite book |doi=10.1016/B978-0-12-386931-9.00014-3 |chapter=The Structural Basis for the Allosteric Regulation of Ribonucleotide Reductase |title=Oligomerization in Health and Disease |series=Progress in Molecular Biology and Translational Science |year=2013 | vauthors = Ahmad MF, Dealwis CG |volume=117 |pages=389–410 |pmid=23663976 |pmc=4059395 |isbn=9780123869319 }}</ref> When bound to ATP, RNR is active. When ATP or dATP is bound to the S site, RNR will catalyze synthesis of dCDP and dUDP from CDP and UDP. dCDP and dUDP can go on to indirectly make dTTP. dTTP bound to the S site will catalyze synthesis of dGDP from GDP, and binding of dGDP to the S site will promote synthesis of dADP from ADP.<ref>{{cite journal | vauthors = Fairman JW, Wijerathna SR, Ahmad MF, Xu H, Nakano R, Jha S, Prendergast J, Welin RM, Flodin S, Roos A, Nordlund P, Li Z, Walz T, Dealwis CG | title = Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization | journal = Nature Structural & Molecular Biology | volume = 18 | issue = 3 | pages = 316–22 | date = March 2011 | pmid = 21336276 | doi = 10.1038/nsmb.2007 | pmc=3101628}}</ref> dADP is then phosphorylated to give dATP, which can bind to the A site and turn RNR off.<ref name=":7" /> | ||
| == Other cellular roles == | == Other cellular roles == | ||
| Line 39: | Line 40: | ||
| === ATP as a source of cellular energy === | === ATP as a source of cellular energy === | ||
| ] | ] | ||
| ] is the primary energy currency of the cell.<ref>{{cite web |title=ATP |url=https://www.nature.com/scitable/definition/atp-318/ |website=Scitable }}</ref> Despite being synthesized through the metabolic pathway described above, it is primarily synthesized during both ]<ref>{{cite web |title=Mitochondria, Cell Energy, ATP Synthase |url=https://www.nature.com/scitable/topicpage/mitochondria-14053590/ |website=Scitable }}</ref> and ]<ref>{{Cite web|url=http://plantsinaction.science.uq.edu.au/content/124-atp-synthesis|title=ATP Synthesis |
] is the primary energy currency of the cell.<ref>{{cite web |title=ATP |url=https://www.nature.com/scitable/definition/atp-318/ |website=Scitable }}</ref> Despite being synthesized through the metabolic pathway described above, it is primarily synthesized during both ]<ref>{{cite web |title=Mitochondria, Cell Energy, ATP Synthase |url=https://www.nature.com/scitable/topicpage/mitochondria-14053590/ |website=Scitable }}</ref> and ]<ref>{{Cite web|url=http://plantsinaction.science.uq.edu.au/content/124-atp-synthesis|title=ATP Synthesis|website=Plants in Action|access-date=2017-11-12}}</ref> by ]. ATP synthase couples the synthesis of ATP from ADP and phosphate with an ] generated by the pumping of protons through either the ] (cellular respiration) or the ] (photosynthesis).<ref>{{cite journal | vauthors = Jonckheere AI, Smeitink JA, Rodenburg RJ | title = Mitochondrial ATP synthase: architecture, function and pathology | journal = Journal of Inherited Metabolic Disease | volume = 35 | issue = 2 | pages = 211–25 | date = March 2012 | pmid = 21874297 | pmc = 3278611 | doi = 10.1007/s10545-011-9382-9 }}</ref> This electrochemical gradient is necessary because the formation of ATP is ]. | ||
| The ] to ADP and |
The ] to ADP and P<sub>i</sub> proceeds as follows:<ref>{{cite journal | vauthors = Dittrich M, Hayashi S, Schulten K | title = On the mechanism of ATP hydrolysis in F1-ATPase | journal = Biophysical Journal | volume = 85 | issue = 4 | pages = 2253–66 | date = October 2003 | pmid = 14507690 | pmc = 1303451 | doi = 10.1016/S0006-3495(03)74650-5 | bibcode = 2003BpJ....85.2253D }}</ref> | ||
| :<chem>ATP + H2O -> ADP + |
:<chem>ATP + H2O -> ADP + P_{i}</chem> | ||
| This reaction is ] and releases 30.5 kJ/mol of energy.<ref |
This reaction is ] and releases 30.5 kJ/mol of energy.<ref name=Khan-ATP /> In the cell, this reaction is often coupled with unfavourable reactions to provide the energy for them to proceed.<ref>{{Cite web|url=https://courses.lumenlearning.com/boundless-biology/chapter/atp-adenosine-triphosphate/|title=ATP: Adenosine Triphosphate {{!}} Boundless Biology|website=courses.lumenlearning.com-US|access-date=2017-11-12}}</ref> ] is occasionally used for energy-coupling in a similar manner.<ref>{{cite journal | vauthors = Carvalho AT, Szeler K, Vavitsas K, Åqvist J, Kamerlin SC | title = Modeling the mechanisms of biological GTP hydrolysis | journal = Archives of Biochemistry and Biophysics | volume = 582 | issue = Supplement C | pages = 80–90 | date = September 2015 | pmid = 25731854 | doi = 10.1016/j.abb.2015.02.027 | series = Special issue in computational modeling on biological systems | url = http://urn.kb.se/resolve?urn=urn:nbn:se:uu:diva-264885 | doi-access = free }}</ref> ] | ||
| === GTP signal transduction === | === GTP signal transduction === | ||
| GTP is essential for ], especially with ]s. G proteins are coupled with a cell membrane bound receptor.<ref name=gpcr14047471/> This whole complex is called a ] (GPCR). G proteins can bind either GDP or GTP. When bound to GDP, G proteins are inactive. When a ] binds a GPCR, an ] change in the G protein is triggered, causing GDP to leave and be replaced by GTP.<ref name=":3">{{Cite |
GTP is essential for ], especially with ]s. G proteins are coupled with a cell membrane bound receptor.<ref name=gpcr14047471/> This whole complex is called a ] (GPCR). G proteins can bind either GDP or GTP. When bound to GDP, G proteins are inactive. When a ] binds a GPCR, an ] change in the G protein is triggered, causing GDP to leave and be replaced by GTP.<ref name=":3">{{Cite encyclopedia|url=https://www.britannica.com/science/G-protein-coupled-receptor|title=G protein-coupled receptor (GPCR) {{!}} biochemistry|encyclopedia=Encyclopedia Britannica|access-date=2017-11-12}}</ref> GTP activates the alpha subunit of the G protein, causing it to dissociate from the G protein and act as a downstream effector.<ref name=":3" /> | ||
| == Nucleoside analogues == | == Nucleoside analogues == | ||
| ]s can be used to treat ].<ref name=":4">{{Cite web|url=http://www.mdpi.com/journal/molecules/special_issues/nucleoside_analogues|title=Nucleoside Analogues |
]s can be used to treat ].<ref name=":4">{{Cite web|url=http://www.mdpi.com/journal/molecules/special_issues/nucleoside_analogues|title=Nucleoside Analogues|website=Molecules|access-date=2017-11-13}}</ref> Nucleoside analogues are nucleosides that are structurally similar (analogous) to the nucleosides used in DNA and RNA synthesis.<ref>{{cite journal | vauthors = Jordheim LP, Durantel D, Zoulim F, Dumontet C | title = Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases | journal = Nature Reviews. Drug Discovery | volume = 12 | issue = 6 | pages = 447–64 | date = June 2013 | pmid = 23722347 | doi = 10.1038/nrd4010 | s2cid = 39842610 }}</ref> Once these nucleoside analogues enter a cell, they can become ] by a viral enzyme. The resulting nucleotides are similar enough to the nucleotides used in DNA or RNA synthesis to be incorporated into growing DNA or RNA strands, but they do not have an available 3' OH group to attach the next nucleotide, causing ].<ref>{{cite journal | vauthors = Ewald B, Sampath D, Plunkett W | title = Nucleoside analogs: molecular mechanisms signaling cell death | journal = Oncogene | volume = 27 | issue = 50 | pages = 6522–37 | date = October 2008 | pmid = 18955977 | doi = 10.1038/onc.2008.316 | s2cid = 23817516 | doi-access = }}</ref> This can be exploited for therapeutic uses in viral infections because viral DNA polymerase recognizes certain nucleotide analogues more readily than eukaryotic DNA polymerase.<ref name=":4" /> For example, ] is used in the treatment of ].<ref name=":10" /> Some less selective nucleoside analogues can be used as ] agents to treat cancer,<ref>{{cite journal | vauthors = Galmarini CM, Mackey JR, Dumontet C | title = Nucleoside analogues and nucleobases in cancer treatment | journal = The Lancet. Oncology | volume = 3 | issue = 7 | pages = 415–24 | date = July 2002 | pmid = 12142171 | doi=10.1016/s1470-2045(02)00788-x}}</ref> such as ] (ara-C) in the treatment of certain forms of ].<ref name="Galmarini_2001" /> | ||
| Resistance to nucleoside analogues is common, and is frequently due to a mutation in the enzyme that phosphorylates the nucleoside after entry into the cell.<ref name="Galmarini_2001">{{cite journal | vauthors = Galmarini CM, Mackey JR, Dumontet C | title = Nucleoside analogues: mechanisms of drug resistance and reversal strategies | journal = Leukemia | volume = 15 | issue = 6 | pages = 875–90 | year = 2001 | pmid = 11417472 | doi = 10.1038/sj.leu.2402114 | doi-access = |
Resistance to nucleoside analogues is common, and is frequently due to a mutation in the enzyme that phosphorylates the nucleoside after entry into the cell.<ref name="Galmarini_2001">{{cite journal | vauthors = Galmarini CM, Mackey JR, Dumontet C | title = Nucleoside analogues: mechanisms of drug resistance and reversal strategies | journal = Leukemia | volume = 15 | issue = 6 | pages = 875–90 | year = 2001 | pmid = 11417472 | doi = 10.1038/sj.leu.2402114 | s2cid = 760764 | doi-access = }}</ref> This is common in nucleoside analogues used to treat HIV/AIDS.<ref>{{cite journal | vauthors = Menéndez-Arias L | title = Mechanisms of resistance to nucleoside analogue inhibitors of HIV-1 reverse transcriptase | journal = Virus Research | volume = 134 | issue = 1–2 | pages = 124–46 | date = June 2008 | pmid = 18272247 | doi = 10.1016/j.virusres.2007.12.015 }}</ref> | ||
| == See also == | == See also == | ||
Latest revision as of 14:29, 29 October 2024
Class of chemical compoundsA nucleoside triphosphate is a nucleoside containing a nitrogenous base bound to a 5-carbon sugar (either ribose or deoxyribose), with three phosphate groups bound to the sugar. They are the molecular precursors of both DNA and RNA, which are chains of nucleotides made through the processes of DNA replication and transcription. Nucleoside triphosphates also serve as a source of energy for cellular reactions and are involved in signalling pathways.
Nucleoside triphosphates cannot easily cross the cell membrane, so they are typically synthesized within the cell. Synthesis pathways differ depending on the specific nucleoside triphosphate being made, but given the many important roles of nucleoside triphosphates, synthesis is tightly regulated in all cases. Nucleoside analogues may also be used to treat viral infections. For example, azidothymidine (AZT) is a nucleoside analogue used to prevent and treat HIV/AIDS.
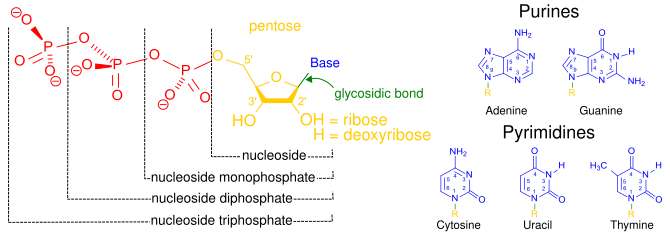
Naming
The term nucleoside refers to a nitrogenous base linked to a 5-carbon sugar (either ribose or deoxyribose). Nucleotides are nucleosides covalently linked to one or more phosphate groups. To provide information about the number of phosphates, nucleotides may instead be referred to as nucleoside (mono, di, or tri) phosphates. Thus, nucleoside triphosphates are a type of nucleotide.
Nucleotides are commonly abbreviated with 3 letters (4 or 5 in case of deoxy- or dideoxy-nucleotides). The first letter indicates the identity of the nitrogenous base (e.g., A for adenine, G for guanine), the second letter indicates the number of phosphates (mono, di, tri), and the third letter is P, standing for phosphate. Nucleoside triphosphates that contain ribose as the sugar are conventionally abbreviated as NTPs, while nucleoside triphosphates containing deoxyribose as the sugar are abbreviated as dNTPs. For example, dATP stands for deoxyribose adenosine triphosphate. NTPs are the building blocks of RNA, and dNTPs are the building blocks of DNA.
The carbons of the sugar in a nucleoside triphosphate are numbered around the carbon ring starting from the original carbonyl of the sugar. Conventionally, the carbon numbers in a sugar are followed by the prime symbol (‘) to distinguish them from the carbons of the nitrogenous base. The nitrogenous base is linked to the 1’ carbon through a glycosidic bond, and the phosphate groups are covalently linked to the 5’ carbon. The first phosphate group linked to the sugar is termed the α-phosphate, the second is the β-phosphate, and the third is the γ-phosphate; these are linked to one another by two phosphoanhydride bonds.
DNA and RNA synthesis

The cellular processes of DNA replication and transcription involve DNA and RNA synthesis, respectively. DNA synthesis uses dNTPs as substrates, while RNA synthesis uses rNTPs as substrates. NTPs cannot be converted directly to dNTPs. DNA contains four different nitrogenous bases: adenine, guanine, cytosine and thymine. RNA also contains adenine, guanine, and cytosine, but replaces thymine with uracil. Thus, DNA synthesis requires dATP, dGTP, dCTP, and dTTP as substrates, while RNA synthesis requires ATP, GTP, CTP, and UTP.
Nucleic acid synthesis is catalyzed by either DNA polymerase or RNA polymerase for DNA and RNA synthesis respectively. These enzymes covalently link the free -OH group on the 3’ carbon of a growing chain of nucleotides to the α-phosphate on the 5’ carbon of the next (d)NTP, releasing the β- and γ-phosphate groups as pyrophosphate (PPi). This results in a phosphodiester linkage between the two (d)NTPs. The release of PPi provides the energy necessary for the reaction to occur. It is important to note that nucleic acid synthesis occurs exclusively in the 5’ to 3’ direction.
Nucleoside triphosphate metabolism
Given their importance in the cell, the synthesis and degradation of nucleoside triphosphates is under tight control. This section focuses on nucleoside triphosphate metabolism in humans, but the process is fairly conserved among species. Nucleoside triphosphates cannot be absorbed well, so all nucleoside triphosphates are typically made de novo. The synthesis of ATP and GTP (purines) differs from the synthesis of CTP, TTP, and UTP (pyrimidines). Both purine and pyrimidine synthesis use phosphoribosyl pyrophosphate (PRPP) as a starting molecule.
The conversion of NTPs to dNTPs can only be done in the diphosphate form. Typically a NTP has one phosphate removed to become a NDP, then is converted to a dNDP by an enzyme called ribonucleotide reductase, then a phosphate is added back to give a dNTP.
Purine synthesis
A nitrogenous base called hypoxanthine is assembled directly onto PRPP. This results in a nucleotide called inosine monophosphate (IMP). IMP is then converted to either a precursor to AMP or GMP. Once AMP or GMP are formed, they can be phosphorylated by ATP to their diphosphate and triphosphate forms.
Purine synthesis is regulated by the allosteric inhibition of IMP formation by the adenine or guanine nucleotides. AMP and GMP also competitively inhibit the formation of their precursors from IMP.
Pyrimidine synthesis
A nitrogenous base called orotate is synthesized independently of PRPP. After orotate is made it is covalently attached to PRPP. This results in a nucleotide called orotate monophosphate (OMP). OMP is converted to UMP, which can then be phosphorylated by ATP to UDP and UTP. UTP can then be converted to CTP by a deamination reaction. TTP is not a substrate for nucleic acid synthesis, so it is not synthesized in the cell. Instead, dTTP is made indirectly from either dUDP or dCDP after conversion to their respective deoxyribose forms.
Pyrimidine synthesis is regulated by the allosteric inhibition of orotate synthesis by UDP and UTP. PRPP and ATP are also allosteric activators of orotate synthesis.
Ribonucleotide reductase
Ribonucleotide reductase (RNR) is the enzyme responsible for converting NTPs to dNTPs. Given that dNTPs are used in DNA replication, the activity of RNR is tightly regulated. It is important to note that RNR can only process NDPs, so NTPs are first dephosphorylated to NDPs before conversion to dNDPs. dNDPs are then typically re-phosphorylated. RNR has 2 subunits and 3 sites: the catalytic site, activity (A) site, and specificity (S) site. The catalytic site is where the NDP to dNDP reaction takes place, the activity site determines whether or not the enzyme is active, and the specificity site determines which reaction takes place in the catalytic site.
The activity site can bind either ATP or dATP. When bound to ATP, RNR is active. When ATP or dATP is bound to the S site, RNR will catalyze synthesis of dCDP and dUDP from CDP and UDP. dCDP and dUDP can go on to indirectly make dTTP. dTTP bound to the S site will catalyze synthesis of dGDP from GDP, and binding of dGDP to the S site will promote synthesis of dADP from ADP. dADP is then phosphorylated to give dATP, which can bind to the A site and turn RNR off.
Other cellular roles
ATP as a source of cellular energy

ATP is the primary energy currency of the cell. Despite being synthesized through the metabolic pathway described above, it is primarily synthesized during both cellular respiration and photosynthesis by ATP synthase. ATP synthase couples the synthesis of ATP from ADP and phosphate with an electrochemical gradient generated by the pumping of protons through either the inner mitochondrial membrane (cellular respiration) or the thylakoid membrane (photosynthesis). This electrochemical gradient is necessary because the formation of ATP is energetically unfavourable.
The hydrolysis of ATP to ADP and Pi proceeds as follows:
This reaction is energetically favourable and releases 30.5 kJ/mol of energy. In the cell, this reaction is often coupled with unfavourable reactions to provide the energy for them to proceed. GTP is occasionally used for energy-coupling in a similar manner.

GTP signal transduction
GTP is essential for signal transduction, especially with G proteins. G proteins are coupled with a cell membrane bound receptor. This whole complex is called a G protein-coupled receptor (GPCR). G proteins can bind either GDP or GTP. When bound to GDP, G proteins are inactive. When a ligand binds a GPCR, an allosteric change in the G protein is triggered, causing GDP to leave and be replaced by GTP. GTP activates the alpha subunit of the G protein, causing it to dissociate from the G protein and act as a downstream effector.
Nucleoside analogues
Nucleoside analogues can be used to treat viral infections. Nucleoside analogues are nucleosides that are structurally similar (analogous) to the nucleosides used in DNA and RNA synthesis. Once these nucleoside analogues enter a cell, they can become phosphorylated by a viral enzyme. The resulting nucleotides are similar enough to the nucleotides used in DNA or RNA synthesis to be incorporated into growing DNA or RNA strands, but they do not have an available 3' OH group to attach the next nucleotide, causing chain termination. This can be exploited for therapeutic uses in viral infections because viral DNA polymerase recognizes certain nucleotide analogues more readily than eukaryotic DNA polymerase. For example, azidothymidine is used in the treatment of HIV/AIDS. Some less selective nucleoside analogues can be used as chemotherapy agents to treat cancer, such as cytosine arabinose (ara-C) in the treatment of certain forms of leukemia.
Resistance to nucleoside analogues is common, and is frequently due to a mutation in the enzyme that phosphorylates the nucleoside after entry into the cell. This is common in nucleoside analogues used to treat HIV/AIDS.
See also
References
- ^ "Nucleotides and Bases - Genetics Generation". Genetics Generation. Retrieved 11 November 2017.
- ^ Chargaff E (2012-12-02). The Nucleic Acids. Elsevier. ISBN 9780323144773.
- ^ "Overview of ATP Hydrolysis". Khan Academy. Archived from the original on 2017-12-01. Retrieved 2017-11-11.
- ^ "GPCR". Scitable. 2014.
- "Eating DNA: Dietary Nucleotides in Nutrition". The call of the Honeyguide. 2014-04-09. Retrieved 11 November 2017.
- ^ Wyngaarden JB (1976). "Regulation of purine biosynthesis and turnover". Advances in Enzyme Regulation. 14: 25–42. doi:10.1016/0065-2571(76)90006-6. PMID 184697.
- ^ Galmarini CM, Mackey JR, Dumontet C (2001). "Nucleoside analogues: mechanisms of drug resistance and reversal strategies". Leukemia. 15 (6): 875–90. doi:10.1038/sj.leu.2402114. PMID 11417472. S2CID 760764.
- ^ "Zidovudine Monograph for Professionals - Drugs.com". Drugs.com. Retrieved 30 November 2017.
- Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J (2000). Structure of Nucleic Acids.
- ^ Secrist JA (May 2001). "Nucleoside and nucleotide nomenclature" (PDF). Current Protocols in Nucleic Acid Chemistry. Appendix 1: A.1D.1–A.1D.3. doi:10.1002/0471142700.nca01ds00. hdl:2027.42/143595. PMID 18428808. S2CID 205152902.
- "Nomenclature of Nucleosides". www.biochem.uthscsa.edu. Retrieved 2017-11-11.
- "From DNA to RNA to protein, how does it work?". Science Explained. Retrieved 11 November 2017.
- "Numbering convention for nucleotides". www.biosyn.com. Retrieved 2017-11-11.
- "SparkNotes: DNA Replication and Repair: The Chemistry of the Addition of Substrates of DNA Replication". www.sparknotes.com. Retrieved 2017-11-11.
- "Do You Know the Differences Between DNA and RNA?". ThoughtCo. Retrieved 2017-11-11.
- "Difference Between DNA Polymerase and RNA Polymerase". 2011-12-24. Retrieved 2017-11-11.
- ^ Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J (2000). Nucleic Acid Synthesis.
- Samant S, Lee H, Ghassemi M, Chen J, Cook JL, Mankin AS, Neyfakh AA (February 2008). "Nucleotide biosynthesis is critical for growth of bacteria in human blood". PLOS Pathogens. 4 (2): e37. doi:10.1371/journal.ppat.0040037. PMC 2242838. PMID 18282099.
- Berg JM, Tymoczko JL, Stryer L (2002). Nucleotide Biosynthesis.
- ^ "Nucleotide Metabolism: Nucleic Acid Synthesis". themedicalbiochemistrypage.org. Retrieved 2017-11-15.
- Stubbe J (1990). "Ribonucleotide reductases: amazing and confusing" (PDF). The Journal of Biological Chemistry. 265 (10): 5329–32. doi:10.1016/S0021-9258(19)39357-3. PMID 2180924.
- Berg J, Tymoczko JL, Stryer L (2002). Purine Bases Can Be Synthesized de Novo or Recycled by Salvage Pathways.
- "Purine Synthesis : Synthesis of Purine RiboNucleotides". BiochemDen.com. 2016-03-16. Retrieved 15 November 2017.
- Berg JM, Tymoczko JL, Stryer L (2002). Key Steps in Nucleotide Biosynthesis Are Regulated by Feedback Inhibition.
- ^ Nierlich DP, Magasanik B (1965). "Regulation of purine ribonucleotide synthesis by end product inhibition. the effect of adenine and guanine ribonucleotides on the 5'-phosphoribosyl-pyrophosphate amidotransferase of aerobacter aerogenes". The Journal of Biological Chemistry. 240: 358–65. doi:10.1016/S0021-9258(18)97657-X. PMID 14253438.
- Moffatt BA, Ashihara H (April 2002). "Purine and pyrimidine nucleotide synthesis and metabolism". The Arabidopsis Book. 1: e0018. doi:10.1199/tab.0018. PMC 3243375. PMID 22303196.
- "Pyrimidine Metabolism". www.cliffsnotes.com. Retrieved 2017-11-15.
- Lane AN, Fan TW (February 2015). "Regulation of mammalian nucleotide metabolism and biosynthesis". Nucleic Acids Research. 43 (4): 2466–85. doi:10.1093/nar/gkv047. PMC 4344498. PMID 25628363.
- ^ Kolberg M, Strand KR, Graff P, Andersson KK (June 2004). "Structure, function, and mechanism of ribonucleotide reductases". Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 1699 (1–2): 1–34. doi:10.1016/j.bbapap.2004.02.007. PMID 15158709.
- ^ Ahmad MF, Dealwis CG (2013). "The Structural Basis for the Allosteric Regulation of Ribonucleotide Reductase". Oligomerization in Health and Disease. Progress in Molecular Biology and Translational Science. Vol. 117. pp. 389–410. doi:10.1016/B978-0-12-386931-9.00014-3. ISBN 9780123869319. PMC 4059395. PMID 23663976.
- Fairman JW, Wijerathna SR, Ahmad MF, Xu H, Nakano R, Jha S, Prendergast J, Welin RM, Flodin S, Roos A, Nordlund P, Li Z, Walz T, Dealwis CG (March 2011). "Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization". Nature Structural & Molecular Biology. 18 (3): 316–22. doi:10.1038/nsmb.2007. PMC 3101628. PMID 21336276.
- "ATP". Scitable.
- "Mitochondria, Cell Energy, ATP Synthase". Scitable.
- "ATP Synthesis". Plants in Action. Retrieved 2017-11-12.
- Jonckheere AI, Smeitink JA, Rodenburg RJ (March 2012). "Mitochondrial ATP synthase: architecture, function and pathology". Journal of Inherited Metabolic Disease. 35 (2): 211–25. doi:10.1007/s10545-011-9382-9. PMC 3278611. PMID 21874297.
- Dittrich M, Hayashi S, Schulten K (October 2003). "On the mechanism of ATP hydrolysis in F1-ATPase". Biophysical Journal. 85 (4): 2253–66. Bibcode:2003BpJ....85.2253D. doi:10.1016/S0006-3495(03)74650-5. PMC 1303451. PMID 14507690.
- "ATP: Adenosine Triphosphate | Boundless Biology". courses.lumenlearning.com-US. Retrieved 2017-11-12.
- Carvalho AT, Szeler K, Vavitsas K, Åqvist J, Kamerlin SC (September 2015). "Modeling the mechanisms of biological GTP hydrolysis". Archives of Biochemistry and Biophysics. Special issue in computational modeling on biological systems. 582 (Supplement C): 80–90. doi:10.1016/j.abb.2015.02.027. PMID 25731854.
- ^ "G protein-coupled receptor (GPCR) | biochemistry". Encyclopedia Britannica. Retrieved 2017-11-12.
- ^ "Nucleoside Analogues". Molecules. Retrieved 2017-11-13.
- Jordheim LP, Durantel D, Zoulim F, Dumontet C (June 2013). "Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases". Nature Reviews. Drug Discovery. 12 (6): 447–64. doi:10.1038/nrd4010. PMID 23722347. S2CID 39842610.
- Ewald B, Sampath D, Plunkett W (October 2008). "Nucleoside analogs: molecular mechanisms signaling cell death". Oncogene. 27 (50): 6522–37. doi:10.1038/onc.2008.316. PMID 18955977. S2CID 23817516.
- Galmarini CM, Mackey JR, Dumontet C (July 2002). "Nucleoside analogues and nucleobases in cancer treatment". The Lancet. Oncology. 3 (7): 415–24. doi:10.1016/s1470-2045(02)00788-x. PMID 12142171.
- Menéndez-Arias L (June 2008). "Mechanisms of resistance to nucleoside analogue inhibitors of HIV-1 reverse transcriptase". Virus Research. 134 (1–2): 124–46. doi:10.1016/j.virusres.2007.12.015. PMID 18272247.
| Nucleic acid constituents | |||||||
|---|---|---|---|---|---|---|---|
| Nucleobase | |||||||
| Nucleoside |
| ||||||
| Nucleotide (Nucleoside monophosphate) |
| ||||||
| Nucleoside diphosphate | |||||||
| Nucleoside triphosphate | |||||||
