| Revision as of 20:04, 22 November 2006 view sourceClockworkSoul (talk | contribs)Extended confirmed users, New page reviewers18,824 edits The {{unreferenced}} banner is no longer appropriate: removing.← Previous edit | Revision as of 20:31, 22 November 2006 view source 202.37.227.112 (talk) →HistoryNext edit → | ||
| Line 44: | Line 44: | ||
| The term "bacteria" has traditionally been generally applied to all microscopic, single-celled prokaryotes. Although this term remains in everyday use, the scientific nomenclature changed after the discovery that prokaryotic life actually consists of two very different lines of ] (''see'' ]). Originally called Eubacteria and Archaebacteria, these evolutionary domains are now called Bacteria and ]. | The term "bacteria" has traditionally been generally applied to all microscopic, single-celled prokaryotes. Although this term remains in everyday use, the scientific nomenclature changed after the discovery that prokaryotic life actually consists of two very different lines of ] (''see'' ]). Originally called Eubacteria and Archaebacteria, these evolutionary domains are now called Bacteria and ]. | ||
| == History == | |||
| The first bacteria were observed by ] in 1674 using a single-lens ] of his own design. The name ''bacterium'' was introduced much later, by ] in 1828, derived from the ] βακτηριον meaning "small stick". Because of the difficulty in describing individual bacteria and the importance of their discovery to fields such as medicine, biochemistry, and geochemistry, the history of bacteriology is generally described as the history of ]. | |||
| ==Morphology and Associations== | ==Morphology and Associations== | ||
Revision as of 20:31, 22 November 2006
Bacteria (singular: bacterium) are microscopic, unicellular organisms. They are often spherical (with the suffix -coccus), rod-shaped (with the suffix -bacillus) or spiral-shaped. They are 0.5-5 µm in the longest dimension, although the wide diversity of bacteria can display a huge variety of morphologies. The study of bacteria is known as bacteriology, a branch of microbiology.
Bacteria are ubiquitous in the environment, living in every possible habitat on the planet including, but by no means limited to, soil, underwater, deep in the earth's crust, and even such environments as sulfuric acid and nuclear waste. There are typically ten billion bacterial cells in a gram of soil, and one hundred thousand bacterial cells in a millilitre of sea water. In all, there may be around five million trillion trillion (5 × 10) bacteria in the world. Bacteria play an important role in the cycling of nutrients in the environment, and many important steps in the nutrient cycle are catalysed exclusively by bacteria, such as the fixation of nitrogen from the atmosphere.
There are more bacterial cells on each of our bodies than there are cells of our own and bacteria are a natural component of the human body, particularly on the skin and in the mouth and intestinal tract. However, pathogenic bacteria cause many infectious diseases, including cholera, leprosy and bubonic plague. The most common bacterial disease is tuberculosis, which kills about 2 million people every year, mostly in sub-Saharan Africa. In developed countries, the use of antibiotics allows the treatment of many bacterial infections, but antibiotic resistance is becoming increasingly common.
Bacteria are also important to numerous industrial processes, such as wastewater treatment, the production of cheese and yoghurt and more recently, the industrial production of antibiotics and other chemicals.
The term "bacteria" has traditionally been generally applied to all microscopic, single-celled prokaryotes. Although this term remains in everyday use, the scientific nomenclature changed after the discovery that prokaryotic life actually consists of two very different lines of evolution (see three-domain system). Originally called Eubacteria and Archaebacteria, these evolutionary domains are now called Bacteria and Archaea.
Morphology and Associations
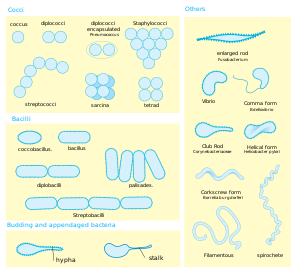
Bacteria display a wide diversity of shapes and sizes, called morphologies. Bacterial cells are typically 0.5-5 μm in length, however some species, for example Thiomargarita namibiensis and Epulopiscium fishelsoni, may be up to 500 µm (0.5 mm) long and are visible to the unaided eye. Among the smallest bacteria are members of the genus Mycoplasma which measure only 0.3 µm, as small as some large viruses.
Despite this diversity, each bacterial species tends to display a characteristic morphology. Most bacteria are either spherical, called coccus (pl. cocci, from Greek kókkos, grain, seed) or rod-shaped, called bacillus (pl. baccili, from Latin baculus, stick). Some rod-shaped bacteria, called vibrio, are slightly curved or comma-shaped, while others, called spirilla, form twisted spirals. These shapes are determined by the bacterial cell wall and cytoskeleton.
Many bacterial species exist simply as single cells, while others tend to associate in diploids (pairs), characteristic for example Neisseria, or chains, such as Streptococcus, while members of the genus Staphylococcus, form a characteristic "bunch of grapes" clusters. Bacteria can also be elongated to form filaments, for example the Actinomycetes. Filamentous bacteria are often surrounded by a sheath which contains many individual cells, and certain species, such as the genus Nocardia, form complex, branched filaments, similar in appearance to fungal mycelia.
Despite their apparent simplicity, bacteria can also form more complex associations. Bacteria can attach to surfaces and form dense aggregations or biofilms. Bacteria living in biofilms can display a complex arrangement of cells and extracellular components, forming secondary structures such as microcolonies, through which there are networks of channels to enable better diffusion of nutrients. In natural environments a majority of bacteria are bound to surfaces in biofilms.
Other morphological changes are possible, for example, under nutrient starvation, Myxobacteria aggregate and form multicellular fruiting bodies which can be up to 500 µm long and contain in the order of 100,000 cells. Cells in these fruiting bodies differentiate into a dormant state to become myxospores, which are more resistant to desiccation and other adverse environmental conditions than the ordinary cells.
Certain bacteria form close spatial associations that are essential for their survival. One such association, called interspecies hydrogen transfer, occurs between clusters of anaerobic bacteria that consume organic acids and produce hydrogen, and methanogenic Archaea that consume hydrogen. These bacteria are unable to consume the organic acids and grow when hydrogen accumulates in their surroundings, and only the intimate association with the hydrogen-consuming Archaea can keep the hydrogen concentration low enough to allow them to grow.
In some instances bacteria have become so closely associated with another cell that they are actually inside of another cell. Such intracellular symbioses are the origin of the mitochondria and chloroplasts, which are descended from bacteria that entered into symbiotic associations with eukaryotic cells early in the evolution of life. Chlamydia are a phylum of bacteria that have evolved such that they can grow and reproduce only as symbionts in the cells of other organisms.
Cellular structure

Intracellular structures
The bacterial cell is bound by a lipid membrane, or plasma membrane, which encompasses the contents of the cell, or cytoplasm, and acts as a barrier that holds nutrients, proteins and other essential molecules within the cell. Bacteria do not have membrane-bound organelles in the cytoplasm and thus contain few intracellular structures (see prokaryotes). They lack mitochondria, chloroplasts and the other organelles present in eukaryotic cells, such as the golgi apparatus and endoplasmic reticulum.
Many important biochemical reactions, such as energy generation, occur due to concentration gradients of certain molecules across membranes that create a potential difference, analogous to a battery. The absence of internal membranes in bacteria means that these reactions, such as electron transport occur across the plasma membrane, between the cytoplasm and the periplasmic space.
Bacteria do not have a membrane-bound nucleus and their genetic material is typically a single chromosome located in the cytoplasm as an irregularly-shaped body called the nucleoid. The nucleoid consists mainly of the chromosome but has also associated proteins and RNA. Like all living organisms bacteria contain ribosomes for the production of proteins, but the structure of the ribosome is uniquely different from Eukarya and Archaea. The order Planctomycetes are an exception to the general absence of internal membranes in bacteria, as they have a membrane around their nucleoid and contain other membrane-bound cellular structures.
Some bacteria also produce intracellular nutrient storage granules, such as glycogen, polyphosphate, sulfur or polyhydroxyalkanoates. These granules enable bacteria to store compounds for later use. Certain bacterial species, such as the photosynthetic Cyanobacteria, produce internal gas vesicles which they use to regulate their buoyancy to regulate the optimal light intensity or nutrient levels.
Extracellular structures
External to the cell membrane is the bacterial cell wall. Bacterial cell walls are composed of peptidoglycan (called Murein in older sources), and are thus different from the cell walls of plants and fungi which have cell walls of cellulose and chitin, respectively. The cell wall of bacteria is also distinct from that of Archaea, which do not contain peptidoglycan. The cell wall is essential to the survival of bacteria; the antibiotic penicillin is able to effectively kill bacteria by inhibiting a step in the synthesis of peptidoglycan and stopping the production of the cell wall.
There are broadly speaking two different arrangements of the cell wall in bacteria, called Gram positive and Gram negative. The names originate from the reaction of cells to the Gram stain, a test long employed for the laboratory classification of bacterial species.
Gram positive bacteria possess a thick cell wall containing many layers of peptidoglycan and teichoic acids. In contrast, Gram negative bacteria have a relatively thin cell wall consisting of a few layers of peptidoglycan surrounded by an outer lipid membrane containing lipopolysaccharides and lipoproteins. Most bacteria have the Gram negative cell wall; only organisms from the Phyla Firmicutes and Actinobacteria (previously known as the low G+C and high G+C Gram positive bacteria, respectively) have the alternative gram-positive arrangement.
In many bacteria an S-layer of rigidly-arrayed protein molecules cover the outside of the cell. This layer provides chemical and physical protection for the cell surface and can act as a macromolecular diffusion barrier. S-layers have diverse but mostly poorly-understood functions, but are known to act as virulence factors in Campylobacter spp. and help define cell shape in Thermoproteus spp.
Flagella are rigid protein structures, about 20 nm in diameter and up to 20 µm in length, that are used for motility (see Motility, below). Flagella are driven by the energy released by the transfer of ions down an electrochemical gradient across the cell membrane. Bacterial species differ in the number and arrangement of flagella on their surface; some have a single flagellum (monotrichous), a flagellum at each end (amphitrichous), clusters of flagella at the poles of the cell (lophotrichous), while others have flagella distributed over the entire surface of the cell (peritrichous).
Fimbriae are fine appendages of protein, just 2-10 nm in diameter and up to several micrometers in length. They are distributed over the surface of the cell and resemble fine hairs when the cells are visualised under the electron microscope. Fimbriae are believed to be involved in attachment to solid surfaces or to other cells and are often importance in the virulence of bacterial pathogens. Pili (sing. pilus) are cellular appendages, slightly larger than fimbriae, that enable the transfer of genetic material between bacterial cells, called conjugation (see Bacterial Genetics, below).
Capsules or slime layers are produced by many bacteria to surround their cells and vary in structural complexity; ranging from disorganised slime layers of extra-cellular polymer, to the highly structured capsule or glycocalyx. These structures can protect cells from engulfment by eukaryotic cells such as macrophages, they can act as antigens and be involved in cell recognition, they can also play a role in attachment to surfaces and biofilm formation.
Spores
Certain genera of gram-positive bacteria, such as Bacillus, Clostridium Sporohalobacter, Anaerobacter and Heliobacterium, are capable of forming highly resistant dormant structures called endospores. Endospores have a distinct structure which is different from plant and fungal spores. Endospores show no detectable metabolism and can survive extreme environmental and chemical stresses, such as high levels of UV light, gamma radiation, heat, pressure and dessication. In this dormant state, these extremely tough organisms may remain viable for millions of years. Spores can also be a cause of disease, as deep puncture wounds that are contaminated with soil may allow infection by the bacterium Clostridium tetani, the cause of tetanus. This bacterium produces endospores which can survive in the environment on surfaces like rusty nails and cause lethal infections when the skin is pierced and the endospores enter and germinate.
Metabolism
Main article: Microbial metabolism
In contrast to higher organisms, bacteria exhibit an extremely wide variety of metabolic types. In fact, it is widely accepted that eukaryotic metabolism is largely a derivative of bacterial metabolism with mitochondria having descended from a lineage within the α-Proteobacteria and chloroplasts from the Cyanobacteria by ancient endosymbiotic events. The distribution of metabolic traits within a group of organisms has traditionally been used to define their taxonomy, although these traits often do not correspond with genetic techniques (see groups and identification below).
Bacterial metabolism can be divided broadly on the basis of the kind of energy used for growth, electron donors and electron acceptors and by the source of carbon used. Most bacteria are heterotrophic; using organic carbon compounds as both carbon and energy sources. In aerobic organisms, oxygen is used as the terminal electron acceptor. In anaerobic organisms other inorganic compounds, such as nitrate, sulfate or carbon dioxide as terminal electron acceptors leading to the environmentally important processes of denitrification, sulfate reduction and acetogenesis, respectively. Non-respiratory anaerobes use fermentation to generate energy and reducing power, secreting metabolic by-products (such as ethanol in brewing) as waste. Facultative anaerobes can switch between fermentation and different terminal electron acceptors depending on the environmental conditions in which they find themselves. As an alternative to heterotrophy some bacteria such as cyanobacteria and purple bacteria are autotrophic, meaning that they obtain cellular carbon by fixing carbon dioxide.
Energy metabolism of bacteria is either based on phototrophy, the use of light: or on chemotrophy, the use of chemical substances for fueling life processes. Lithotrophic bacteria use inorganic electron donors for respiration (chemolithotrophs) or biosynthesis and carbon dioxide fixation (photolithotrophs). In contrast, organotrophs use organic compounds as electron donors for biosynthetic reactions. Common inorganic electron donors are hydrogen, carbon monoxide, ammonia (leading to nitrification), ferrous iron and other reduced metal ions, and several reduced sulfur compounds. Unusually, the gas methane can be used by methanotrophic bacteria as both a source of electrons and a substrate for carbon anabolism. In both aerobic phototrophy and chemolithotrophy oxygen is used as a terminal electron acceptor, while under anaerobic conditions inorganic compounds (see above) are used instead. Most photolithotrophic and chemolithotrophic organisms are autotrophic, whereas photoorganotrophic and chemoorganotrophic organisms are heterotrophic.
In addition to carbon, some bacteria also fix nitrogen gas (nitrogen fixation) using the enzyme nitrogenase. This environmentally important trait can be found in bacteria of nearly all the metabolic types listed above, but is not universal.
Growth and reproduction
All bacteria reproduce through asexual reproduction (one parent) binary fission, under optimal conditions bacteria can grow extremely rapidly and can double as quickly as every 9.8 minutes. Two identical clone daughter cells are produced. Some bacteria, while still reproducing asexually, form more complex reproductive structures that facilitate the dispersal of the newly-formed daughter cells. Examples include fruiting body formation by Myxococcus and arial hyphae formation by Streptomyces, or budding. Budding is resulted of a 'bud' of a cell growing from another cell, and then finally breaking away.

Only about half of the known bacterial phyla have species that can be cultured in the laboratory and our knowledge of bacteria as a whole is biased towards those organisms that can be isolated in this way. In the laboratory, bacteria are usually grown using solid or liquid media. Solid growth media such as agar plates are used to isolate pure cultures of a bacterial strain. However, when measurement of growth or large volumes of cells are required, liquid growth media are used. Growth in stirred liquid media occurs as an even cell suspension, making the cultures easy to divide and transfer compared to solid media, although isolating single bacteria from liquid media is extremely difficult.
Most laboratory techniques for growing bacteria use high levels of nutrients to produce large amounts of cells cheaply and quickly. However, in natural environments nutrients are limited, meaning that bacteria cannot continue to reproduce indefinitely. This constant limitation of nutrients has led the evolution of different growth strategies in different types of organisms (see R/K selection theory). Some possess the ability to grow extremely rapidly when nutrients become available, such as the formation of algal (and cyanobacterial) blooms that often occur in lakes during the summer. Other organisms have devised more specialized strategies to make them more successful in a harsh environment, such as the production of multiple antibiotics by Streptomyces; that inhibit the growth of competing microorganisms. In a natural environment, many organisms live in communities (e.g. biofilms) which may allow for increased supply of nutrients and protection of environmental stresses. These relationships can be essential for growth of a particular organism or group of organisms (syntrophy).
Bacterial growth follows three distinct phases. When an old stock of bacteria is diluted into fresh media these cells need to adapt to the nutrient rich environment. The first phase of growth is the lag phase, a period of slow growth when the cells are adapting to fast growth. The lag phase has high biosynthesis rates, as enzymes needed to metabolise a variety of substrates are produced. The second phase of growth is the logarithmic phase (log phase), also known as the exponential phase. The log phase is marked by rapid exponential growth. The rate at which cells grow during this phase is known as the growth rate (k) and the time it takes the cells to double is known as the generation time (g). During the log phase, nutrients are metabolised at maximum speed until one of the nutrients is depleted and starts limiting growth. The final phase of growth is the stationary phase and is caused by depleted nutrients. The cells begin to reduce their metabolic activity, as well consume their own non-essential proteins. The stationary phase is a transition from rapid growth to a stress response state and there is increased expression of genes involved in DNA repair, antioxidant metabolism and nutrient transport. The limitations of this discontinuous growth pattern can be avoided by the use of a chemostat, which maintains a bacterial culture under steady-state conditions by continuously adding nutrients and removing waste products and cells. Large chemostats are often used for industrial-scale microbial processes.
Genetic variation
Bacteria, as asexual organisms, inherit an identical copy of their parent's genes (i.e. are clonal). All bacteria, however, have the ability to evolve through selection on changes to their genetic material (DNA) which arise either through mutation or genetic recombination. Mutation occurs as a result of errors made during the replication of DNA. It occurs naturally and as a result of the presence of mutagens. Mutation rates can vary among different species of bacteria. The most frequent genetic changes in bacterial genomes come from random mutation. Some bacteria can also undergo genetic recombination. This can occur when bacteria take-up exogenous environmental DNA from closely related genera in a process called transformation. In the process of transduction, a virus can alter the DNA of a bacterium by becoming lysogenic and introducing foreign DNA into the host chromosome, which can then be transcribed and replicated. This gene acquisition from the environment is called horizontal gene transfer and may be common under natural conditions.
Because of their ability to quickly grow, and the relative ease with which they can be manipulated, bacteria have historically been the workhorses for the fields of molecular biology, genetics and biochemistry. By making mutations in bacterial DNA and examining the resulting phenotypes, scientists have been able to determine the function of many different genes and enzymes. Lessons learned from bacteria can then be applied to more complex organisms which are often more difficult to study.
Movement

Motile bacteria can move about, using flagella, bacterial gliding, or changes of buoyancy. A unique group of bacteria, the spirochaetes, have structures similar to flagella, called axial filaments, between two membranes in the periplasmic space. They have a distinctive helical body that twists about as it moves.
Bacterial flagella are arranged in many different ways. Bacteria can have a single polar flagellum at one end of a cell, clusters of many flagella at one end or flagella scattered all over the cell, as with peritrichous. Often in the close vicinity of the flagella a specialized region of the cell membrane the polar membrane can be discerned in ultrathin sections. Many bacteria (such as E. coli) have two distinct modes of movement: forward movement (swimming) and tumbling. The tumbling allows them to reorient and introduces an important element of randomness in their forward movement. (See external links below for link to videos.)
Motile bacteria are attracted or repelled by certain stimuli, behaviors called taxes - for instance, chemotaxis, phototaxis, mechanotaxis, and magnetotaxis. In one peculiar group, the myxobacteria, individual bacteria attract to form swarms and may differentiate to form fruiting bodies. The myxobacteria move only when on solid surfaces, unlike E. coli which is motile in liquid or solid media.
Groups and identification
Historically, bacteria as originally studied by botanists were classified in the same way as plants, that is, mainly by shape. Bacteria come in a variety of different cell morphologies (shapes), including bacillus (rod-shape), coccus (spherical), spirillum (helical), and vibrio (curved bacillus). However, because of their small size bacteria are relatively uniform in shape and therefore classification based on morphology was unsuccessful. The first formal classification scheme was developed following the development of the Gram stain by Hans Christian Gram which separates bacteria based on the structural characteristics of their cell walls. This scheme included:
- Gracilicutes - Gram negative staining bacteria with a second cell membrane
- Firmicutes - Gram positive staining bacteria with a thick peptidoglycan wall
- Mollicutes - Gram negative staining bacteria with no cell wall or second membrane
- Mendosicutes - atypically staining strains now known to belong to the Archaea
Further developments (essentially) based on this scheme included comparisons of bacteria based on differences in cellular metabolism as determined by a wide variety of specific tests. Bacteria were also classified based on differences in cellular chemical compounds such as fatty acids, pigments, and quinones for example. While these schemes allowed for the differentiation between bacterial strains, it was unclear whether these differences represented variation between distinct species or between strains of the same species. It was not until the utilization of genome-based techniques such as guanine cytosine ratio determination, genome-genome hybridization and gene sequencing (in particular the rRNA gene) that microbial taxonomy developed (or at least is developing) into a stable, accurate classification system. It should be noted, however, that due to the existence numerous historical classification schemes and our current poor understanding of microbial diversity, bacterial taxonomy remains a changing and expanding field.
Benefits and dangers
Bacteria are both harmful and useful to the environment and animals, including humans. The role of bacteria in disease and infection is important. Some bacteria act as pathogens and cause diseases such as tetanus, typhoid fever, diphtheria, syphilis, cholera, food-borne illness, leprosy, and tuberculosis(TB). Sepsis, a systemic infectious syndrome characterized by shock and massive vasodilation, or localized infection, can be caused by bacteria such as Streptococcus, Staphylococcus, or many gram-negative bacteria. Some bacterial infections can spread throughout the host's body and become systemic. In plants, bacteria cause leaf spot, fireblight, and wilts. The mode of infection includes contact, air, food, water, and insect-borne microorganisms. The hosts infected with the pathogens may be treated with antibiotics, which can be classified as bacteriocidal and bacteriostatic, which at concentrations that can be reached in bodily fluids either kill bacteria or hamper their growth, respectively. Antiseptic measures may be taken to prevent infection by bacteria, for example, by swabbing skin with alcohol prior to piercing the skin with the needle of a syringe. Sterilization of surgical and dental instruments is done to make them sterile or pathogen-free to prevent contamination and infection by bacteria. Sanitizers and disinfectants are used to kill bacteria or other pathogens to prevent contamination and risk of infection.
In soil, microorganisms which reside in the rhizosphere (a zone that includes the root surface and the soil that adheres to the root after gentle shaking) help in the transformation of molecular dinitrogen gas as their source of nitrogen, converting it to nitrogenous compounds in a process known as nitrogen fixation. This serves to provide an easily absorbable form of nitrogen for many plants, which cannot fix nitrogen themselves. Many other bacteria are found as symbionts in humans and other organisms. For example, the presence of the gut flora in the human large intestine can contribute to gut immunity, synthesise vitamins such as folic acid and biotin, and ferment complex undigestable carbohydrates.
The ability of bacteria to degrade a variety of organic compounds is remarkable. Highly specialized groups of microorganisms play important roles in the mineralization of specific classes of organic compounds. For example, the decomposition of cellulose, which is one of the most abundant constituents of plant tissues, is mainly brought about by aerobic bacteria that belong to the genus Cytophaga. This ability has also been utilized by humans in industry, waste processing, and bioremediation. Bacteria capable of digesting the hydrocarbons in petroleum are often used to clean up oil spills. Some beaches in Prince William Sound were fertilized in an attempt to facilitate the growth of such bacteria after the infamous 1989 Exxon Valdez oil spill. These efforts were effective on beaches that were not too thickly covered in oil.
Bacteria, often in combination with yeasts and molds, are used in the preparation of fermented foods such as cheese, pickles, soy sauce, sauerkraut, vinegar, wine, and yogurt. Using biotechnology techniques, bacteria can be bioengineered for the production of therapeutic drugs, such as insulin, or for the bioremediation of toxic wastes.
"Friendly bacteria" is a term used to refer to those bacteria that offer some benefit to human hosts, such as Lactobacillus species, which convert milk protein to lactic acid in the gut. The presence of such bacterial colonies also inhibits the growth of potentially pathogenic bacteria (usually through competitive exclusion). Other bacteria that are helpful inside the body are many strains of E. coli, which are harmless in healthy individuals and provide Vitamin K.
See also
- Bacterial growth
- Bacteriocin
- Economic importance of bacteria
- Magnetotactic bacteria
- Microorganism
- Nanobacterium
- Transgenic bacteria
References
- University of Georgia Campus News
- Schulz H, Jorgensen B. "Big bacteria". Annu Rev Microbiol. 55: 105–37. PMID 11544351.
- Robertson J, Gomersall M, Gill P. (1975). "Mycoplasma hominis: growth, reproduction, and isolation of small viable cells". J Bacteriol. 124 (2): 1007–18. PMID 1102522.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Cabeen M, Jacobs-Wagner C (2005). "Bacterial cell shape". Nat Rev Microbiol. 3 (8): 601–10. PMID 16012516.
- Douwes K, Schmalzbauer E, Linde H, Reisberger E, Fleischer K, Lehn N, Landthaler M, Vogt T (2003). "Branched filaments no fungus, ovoid bodies no bacteria: Two unusual cases of mycetoma". J Am Acad Dermatol. 49 (2 Suppl Case Reports): S170-3. PMID 12894113.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Donlan R (2002). "Biofilms: microbial life on surfaces". Emerg Infect Dis. 8 (9): 881–90. PMID 12194761.
- Branda S, Vik S, Friedman L, Kolter R (2005). "Biofilms: the matrix revisited". Trends Microbiol. 13 (1): 20–6. PMID 15639628.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Davey M, O'toole G (2000). "Microbial biofilms: from ecology to molecular genetics". Microbiol Mol Biol Rev. 64 (4): 847–67. PMID 11104821.
- Kaiser D. "Signaling in myxobacteria". Annu Rev Microbiol. 58: 75–98. PMID 15487930.
- Stams A, de Bok F, Plugge C, van Eekert M, Dolfing J, Schraa G (2006). "Exocellular electron transfer in anaerobic microbial communities". Environ Microbiol. 8 (3): 371–82. PMID 16478444.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Dyall S, Brown M, Johnson P (2004). "Ancient invasions: from endosymbionts to organelles". Science. 304 (5668): 253–7. PMID 15073369.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Harold F (1972). "Conservation and transformation of energy by bacterial membranes". Bacteriol Rev. 36 (2): 172–230. PMID 4261111.
- Thanbichler M, Wang S, Shapiro L (2005). "The bacterial nucleoid: a highly organized and dynamic structure". J Cell Biochem. 96 (3): 506–21. PMID 15988757.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Poehlsgaard J, Douthwaite S (2005). "The bacterial ribosome as a target for antibiotics". Nat Rev Microbiol. 3 (11): 870–81. PMID 16261170.
- Fuerst J (2005). "Intracellular compartmentation in planctomycetes". Annu Rev Microbiol. 59: 299–328. PMID 15910279.
- ^ Koch A (2003). "Bacterial wall as target for attack: past, present, and future research". Clin Microbiol Rev. 16 (4): 673–87. PMID 14557293.
- Hugenholtz P (2002). "Exploring prokaryotic diversity in the genomic era". Genome Biol. 3 (2): REVIEWS0003. PMID 11864374.
- Engelhardt H, Peters J (1998). "Structural research on surface layers: a focus on stability, surface layer homology domains, and surface layer-cell wall interactions". J Struct Biol. 124 (2–3): 276–302. PMID 10049812.
- Beveridge T, Pouwels P, Sára M, Kotiranta A, Lounatmaa K, Kari K, Kerosuo E, Haapasalo M, Egelseer E, Schocher I, Sleytr U, Morelli L, Callegari M, Nomellini J, Bingle W, Smit J, Leibovitz E, Lemaire M, Miras I, Salamitou S, Béguin P, Ohayon H, Gounon P, Matuschek M, Koval S (1997). "Functions of S-layers". FEMS Microbiol Rev. 20 (1–2): 99–149. PMID 9276929.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Kojima S, Blair D. "The bacterial flagellar motor: structure and function of a complex molecular machine". Int Rev Cytol. 233: 93–134. PMID 15037363.
- Beachey E (1981). "Bacterial adherence: adhesin-receptor interactions mediating the attachment of bacteria to mucosal surface". J Infect Dis. 143 (3): 325–45. PMID 7014727.
- Silverman P (1997). "Towards a structural biology of bacterial conjugation". Mol Microbiol. 23 (3): 423–9. PMID 9044277.
- Stokes R, Norris-Jones R, Brooks D, Beveridge T, Doxsee D, Thorson L (2004). "The glycan-rich outer layer of the cell wall of Mycobacterium tuberculosis acts as an antiphagocytic capsule limiting the association of the bacterium with macrophages". Infect Immun. 72 (10): 5676–86. PMID 15385466.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Daffé M, Etienne G (1999). "The capsule of Mycobacterium tuberculosis and its implications for pathogenicity". Tuber Lung Dis. 79 (3): 153–69. PMID 10656114.
- Nicholson W, Munakata N, Horneck G, Melosh H, Setlow P (2000). "Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments". Microbiol Mol Biol Rev. 64 (3): 548–72. PMID 10974126.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Nicholson W, Fajardo-Cavazos P, Rebeil R, Slieman T, Riesenman P, Law J, Xue Y (2002). "Bacterial endospores and their significance in stress resistance". Antonie Van Leeuwenhoek. 81 (1–4): 27–32. PMID 12448702.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Vreeland R, Rosenzweig W, Powers D (2000). "Isolation of a 250 million-year-old halotolerant bacterium from a primary salt crystal". Nature. 407 (6806): 897–900. PMID 11057666.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Cano R, Borucki M (1995). "Revival and identification of bacterial spores in 25- to 40-million-year-old Dominican amber". Science. 268 (5213): 1060–4. PMID 7538699.
- Hatheway C (1990). "Toxigenic clostridia". Clin Microbiol Rev. 3 (1): 66–98. PMID 2404569.
- Zumft W (1997). "Cell biology and molecular basis of denitrification". Microbiol Mol Biol Rev. 61 (4): 533–616. PMID 9409151.
- Drake H, Daniel S, Küsel K, Matthies C, Kuhner C, Braus-Stromeyer S (1997). "Acetogenic bacteria: what are the in situ consequences of their diverse metabolic versatilities?". Biofactors. 6 (1): 13–24. PMID 9233536.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Hellingwerf K, Crielaard W, Hoff W, Matthijs H, Mur L, van Rotterdam B (1994). "Photobiology of bacteria". Antonie Van Leeuwenhoek. 65 (4): 331–47. PMID 7832590.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Dalton H (2005). "The Leeuwenhoek Lecture 2000 the natural and unnatural history of methane-oxidizing bacteria" (PDF). Philos Trans R Soc Lond B Biol Sci. 360 (1458): 1207–22. PMID 16147517.
- Zehr J, Jenkins B, Short S, Steward G (2003). "Nitrogenase gene diversity and microbial community structure: a cross-system comparison". Environ Microbiol. 5 (7): 539–54. PMID 12823187.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Eagon R. "Pseudomonas natriegens, a marine bacterium with a generation time of less than 10 minutes". J Bacteriol. 83: 736–7. PMID 13888946.
- Rappé M, Giovannoni S. "The uncultured microbial majority". Annu Rev Microbiol. 57: 369–94. PMID 14527284.
- Paerl H, Fulton R, Moisander P, Dyble J. "Harmful freshwater algal blooms, with an emphasis on cyanobacteria". ScientificWorldJournal. 1: 76–113. PMID 12805693.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Challis G, Hopwood D. "Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species". Proc Natl Acad Sci U S A. 100 Suppl 2: 14555–61. PMID 12970466.
- Kooijman S, Auger P, Poggiale J, Kooi B (2003). "Quantitative steps in symbiogenesis and the evolution of homeostasis". Biol Rev Camb Philos Soc. 78 (3): 435–63. PMID 14558592.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Prats C, López D, Giró A, Ferrer J, Valls J (2006). "Individual-based modelling of bacterial cultures to study the microscopic causes of the lag phase". J Theor Biol. 241 (4): 939–53. PMID 16524598.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Hecker M, Völker U. "General stress response of Bacillus subtilis and other bacteria". Adv Microb Physiol. 44: 35–91. PMID 11407115.
- Wolska K (2003). "Horizontal DNA transfer between bacteria in the environment". Acta Microbiol Pol. 52 (3): 233–43. PMID 14743976.
- Barea J, Pozo M, Azcón R, Azcón-Aguilar C (2005). "Microbial co-operation in the rhizosphere". J Exp Bot. 56 (417): 1761–78. PMID 15911555.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - O'Hara A, Shanahan F (2006). "The gut flora as a forgotten organ". EMBO Rep. 7 (7): 688–93. PMID 16819463.
- Some text in this entry was merged with the Nupedia article entitled Bacteria, written by Nagina Parmar; reviewed and approved by the Biology group (editor: Gaytha Langlois, lead reviewer: Gaytha Langlois, lead copyeditors: Ruth Ifcher and Jan Hogle)
 This article incorporates public domain material from Science Primer. NCBI. Archived from the original on 2009-12-08.
This article incorporates public domain material from Science Primer. NCBI. Archived from the original on 2009-12-08.
Further reading
- Alcamo, I. Edward. Fundamentals of Microbiology. 5th ed. Menlo Park, California: Benjamin Cumming, 1997.
- Atlas, Ronald M. Principles of Microbiology. St. Louis, Missouri: Mosby, 1995.
- Holt, John.G. Bergey's Manual of Determinative Bacteriology. 9th ed. Baltimore, Maryland: Williams and Wilkins, 1994.
- Hugenholtz P, Goebel BM, Pace NR (1998). "Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity". J Bacteriol. 180 (18): 4765–74. PMID 9733676.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Koshland, Daniel E., Jr. (1977). "A Response Regulator Model in a Simple Sensory System". Science. 196: 1055–1063. PMID 870969.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Stanier, R.Y., J. L. Ingraham, M. L. Wheelis, and P. R. Painter. General Microbiology. 5th ed. Upper Saddle River, New Jersey: Prentice Hall, 1986.
External links
- Bacterial Nomenclature Up-To-Date from DSMZ
- Bacterial Growth and Cell Wall (Ger)
- Microminds
- The largest bacteria
- Tree of Life
- Videos of bacteria swimming and tumbling, use of optical tweezers and other fine videos.
- Planet of the Bacteria by Stephen Jay Gould
- Major Groups of Prokaryotes
- Bitter Resistance by Bruce Sterling
- On-line text book on bacteriology
- cyanobacteria in lichens
