| Revision as of 16:54, 29 December 2020 editTorrent97 (talk | contribs)277 edits cleaning up spaces and supplementing a link.Tag: Visual edit← Previous edit | Latest revision as of 12:13, 12 July 2024 edit undoCitation bot (talk | contribs)Bots5,427,476 edits Added bibcode. | Use this bot. Report bugs. | Suggested by Abductive | Category:Carcinogenesis | #UCB_Category 8/44 | ||
| Line 40: | Line 40: | ||
| }} | }} | ||
| '''M<sub>1</sub>G''' ('''pyrimidopurin-10(''3H'')-one''') is a ] which is a ] of ] (BER) of a specific type of ] called M<sub>1</sub>dG. The M<sub>1</sub>dG adduct in turn is formed by a ] between ] nucleotides in ] and either ] (propanedial) <ref name="martnett">{{cite journal | author = Marnett LJ | title = Lipid peroxidation-DNA damage by malondialdehyde | journal = Mutat. Res. | volume = 424 | issue = 1–2 | pages = 83–95 | year = 1999 | pmid = 10064852 | doi = 10.1016/S0027-5107(99)00010-X }}</ref> or ].<ref name="seto">{{cite journal |vauthors=Seto H, Okuda T, Takesue T, Ikemura T | title = Reaction of Malonaldehyde with Nucleic Acid. I. Formation of Fluorescent Pyrimidopurin-10(3H)-one Nucleosides | journal = Bulletin of the Chemical Society of Japan | volume = 56 | issue = 6 | pages = 1799–1802 | year = 1983 | doi = 10.1246/bcsj.56.1799 | doi-access = free }}</ref> If not repaired, these adducts are ]ic and ]ic. | '''M<sub>1</sub>G''' ('''pyrimidopurin-10(''3H'')-one''') is a ] which is a ] of ] (BER) of a specific type of ] called M<sub>1</sub>dG. The M<sub>1</sub>dG adduct in turn is formed by a ] between ] nucleotides in ] and either ] (propanedial) <ref name="martnett">{{cite journal | author = Marnett LJ | title = Lipid peroxidation-DNA damage by malondialdehyde | journal = Mutat. Res. | volume = 424 | issue = 1–2 | pages = 83–95 | year = 1999 | pmid = 10064852 | doi = 10.1016/S0027-5107(99)00010-X | bibcode = 1999MRFMM.424...83M }}</ref> or ].<ref name="seto">{{cite journal |vauthors=Seto H, Okuda T, Takesue T, Ikemura T | title = Reaction of Malonaldehyde with Nucleic Acid. I. Formation of Fluorescent Pyrimidopurin-10(3H)-one Nucleosides | journal = Bulletin of the Chemical Society of Japan | volume = 56 | issue = 6 | pages = 1799–1802 | year = 1983 | doi = 10.1246/bcsj.56.1799 | doi-access = free }}</ref> If not repaired, these adducts are ]ic and ]ic. | ||
| Malondialdehyde is an end product of ]<ref name="seto"/> while ] is a result of DNA peroxidation.<ref name="pmid17311424">{{cite journal |vauthors=Knutson CG, Akingbade D, Crews BC, Voehler M, Stec DF, Marnett LJ | title = Metabolism in vitro and in vivo of the DNA base adduct, M1G | journal = Chem. Res. Toxicol. | volume = 20 | issue = 3 | pages = 550–7 |date=March 2007 | pmid = 17311424 | doi = 10.1021/tx600334x | doi-access = free }}</ref> | Malondialdehyde is an end product of ]<ref name="seto"/> while ] is a result of DNA peroxidation.<ref name="pmid17311424">{{cite journal |vauthors=Knutson CG, Akingbade D, Crews BC, Voehler M, Stec DF, Marnett LJ | title = Metabolism in vitro and in vivo of the DNA base adduct, M1G | journal = Chem. Res. Toxicol. | volume = 20 | issue = 3 | pages = 550–7 |date=March 2007 | pmid = 17311424 | doi = 10.1021/tx600334x | doi-access = free }}</ref> | ||
Latest revision as of 12:13, 12 July 2024
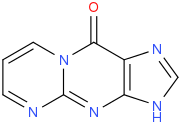
| |
| Names | |
|---|---|
| IUPAC name Pyrimidopurin-10(3H)-one | |
| Identifiers | |
| CAS Number | |
| 3D model (JSmol) | |
| ChemSpider | |
| MeSH | C107643 |
| PubChem CID | |
| UNII | |
| CompTox Dashboard (EPA) | |
InChI
| |
SMILES
| |
| Properties | |
| Chemical formula | C8H5N5O |
| Molar mass | 187.162 g·mol |
| Except where otherwise noted, data are given for materials in their standard state (at 25 °C , 100 kPa).
| |
M1G (pyrimidopurin-10(3H)-one) is a heterocyclic compound which is a by-product of base excision repair (BER) of a specific type of DNA adduct called M1dG. The M1dG adduct in turn is formed by a condensation reaction between guanosine nucleotides in DNA and either malondialdehyde (propanedial) or acrolein. If not repaired, these adducts are mutagenic and carcinogenic.
Malondialdehyde is an end product of lipid peroxidation while acrolein is a result of DNA peroxidation.
M1dG is the major endogenous DNA adduct in humans. M1dG adducts have been detected in cell DNA in liver, leucocytes, pancreas and breast in concentrations of 1-120 per 10 nucleotides. Detection and quantification of M1dG adducts in the body as measured by free M1G is a tool for detecting DNA damage that may lead to cancer. Free M1G is also biomarker for oxidative stress.
References
- ^ Marnett LJ (1999). "Lipid peroxidation-DNA damage by malondialdehyde". Mutat. Res. 424 (1–2): 83–95. Bibcode:1999MRFMM.424...83M. doi:10.1016/S0027-5107(99)00010-X. PMID 10064852.
- ^ Seto H, Okuda T, Takesue T, Ikemura T (1983). "Reaction of Malonaldehyde with Nucleic Acid. I. Formation of Fluorescent Pyrimido[1,2-a]purin-10(3H)-one Nucleosides". Bulletin of the Chemical Society of Japan. 56 (6): 1799–1802. doi:10.1246/bcsj.56.1799.
- Knutson CG, Akingbade D, Crews BC, Voehler M, Stec DF, Marnett LJ (March 2007). "Metabolism in vitro and in vivo of the DNA base adduct, M1G". Chem. Res. Toxicol. 20 (3): 550–7. doi:10.1021/tx600334x. PMID 17311424.
External links
- pyrimido(1,2-a)purin-10(3H)-one at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- 3-(2'-deoxy-beta-D-erythro-pentofuranosyl)pyrimido(1,2-alpha)purin-10(3H)-one at the U.S. National Library of Medicine Medical Subject Headings (MeSH)