A genetically engineered fluorescent protein that changes its fluorescence when bound to the neurotransmitter glutamate. Glutamate-sensitive fluorescent reporters (iGluSnFR, colloquially pronounced 'glue sniffer') are used to monitor the activity of presynaptic terminals by fluorescence microscopy. GluSnFRs are a class of optogenetic sensors used in neuroscience research. In brain tissue, two-photon microscopy is typically used to monitor GluSnFR fluorescence.
Design
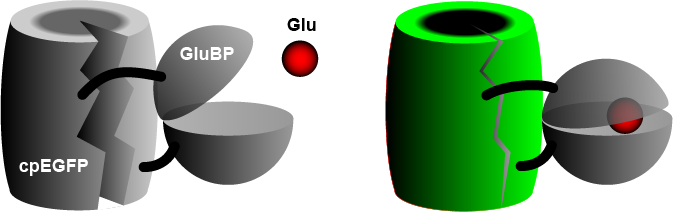
The widely used iGluSnFR consists of a circularly permuted enhanced green fluorescent protein (cpEGFP) fused to a glutamate binding protein (GluBP) from a bacterium. When GluBP binds a glutamate molecule, it changes its shape, pulling the EGFP barrel together, increasing fluorescence. A specific peptide segment (PDGFR) is included to bring the sensor to the outside of the cell membrane. In the more recent version by Aggarwal et al. (2022), researchers introduced iGluSnFR to two additional anchoring domains, a glycosylphostidylinositol (GPI) anchor, and a modified form of the cytosolic -cterminal domain of Stargazin with a PDZ ligand.
History
The first genetically encoded fluorescent glutamate sensors (FLIPE, GluSnFR and SuperGluSnFR) were constructed by attaching cyan-fluorescent protein (CFP) and yellow-fluorescent protein (YFP) to a bacterial glutamate binding protein (GluBP). Glutamate binding changed the distance between CFP and YFP, changing the efficiency of energy transfer (FRET) between the two fluorophores. A breakthrough in visualizing glutamate release was achieved with iGluSnFR, a single-fluorophore glutamate sensor based on EGFP producing a ~5‑fold increase in fluorescence. To measure synaptic transmission at high frequencies, novel iGluSnFR variants with accelerated kinetics have recently been developed.
References
- ^ Aggarwal, Abhi; Liu, Rui; Chen, Yang; Ralowicz, Amelia J.; Bergerson, Samuel J.; Tomaska, Filip; Hanson, Timothy L.; Hasseman, Jeremy P.; Reep, Daniel; Tsegaye, Getahun; Yao, Pantong; Ji, Xiang; Kloos, Marinus; Walpita, Deepika; Patel, Ronak (2022-02-15). "Glutamate indicators with improved activation kinetics and localization for imaging synaptic transmission": 2022.02.13.480251. doi:10.1101/2022.02.13.480251. hdl:20.500.11850/613938.
{{cite journal}}: Cite journal requires|journal=(help) - Hefendehl, J. K.; LeDue, J.; Ko, R. W. Y.; Mahler, J.; Murphy, T. H.; MacVicar, B. A. (2016-11-11). "Mapping synaptic glutamate transporter dysfunction in vivo to regions surrounding Aβ plaques by iGluSnFR two-photon imaging". Nature Communications. 7: 13441. Bibcode:2016NatCo...713441H. doi:10.1038/ncomms13441. PMC 5114608. PMID 27834383.
- ^ Marvin, Jonathan S; Borghuis, Bart G; Tian, Lin; Cichon, Joseph; Harnett, Mark T; Akerboom, Jasper; Gordus, Andrew; Renninger, Sabine L; Chen, Tsai-Wen (2013). "An optimized fluorescent probe for visualizing glutamate neurotransmission". Nature Methods. 10 (2): 162–170. doi:10.1038/nmeth.2333. ISSN 1548-7105. PMC 4469972. PMID 23314171.
- Marvin, Jonathan S.; Schreiter, Eric R.; Echevarría, Ileabett M.; Looger, Loren L. (2011-11-01). "A genetically encoded, high-signal-to-noise maltose sensor". Proteins: Structure, Function, and Bioinformatics. 79 (11): 3025–3036. doi:10.1002/prot.23118. ISSN 1097-0134. PMC 3265398. PMID 21989929.
- Hu, Yonglin; Fan, Cheng-Peng; Fu, Guangsen; Zhu, Deyu; Jin, Qi; Wang, Da-Cheng (2008). "Crystal Structure of a Glutamate/Aspartate Binding Protein Complexed with a Glutamate Molecule: Structural Basis of Ligand Specificity at Atomic Resolution". Journal of Molecular Biology. 382 (1): 99–111. doi:10.1016/j.jmb.2008.06.091. PMID 18640128.
- De Lorimier, Robert M.; Smith, J. Jeff; Dwyer, Mary A.; Looger, Loren L.; Sali, Kevin M.; Paavola, Chad D.; Rizk, Shahir S.; Sadigov, Shamil; Conrad, David W. (2002-11-01). "Construction of a fluorescent biosensor family". Protein Science. 11 (11): 2655–2675. doi:10.1110/ps.021860. ISSN 1469-896X. PMC 2373719. PMID 12381848.
- Okumoto, Sakiko; Looger, Loren L.; Micheva, Kristina D.; Reimer, Richard J.; Smith, Stephen J.; Frommer, Wolf B. (2005-06-14). "Detection of glutamate release from neurons by genetically encoded surface-displayed FRET nanosensors". Proceedings of the National Academy of Sciences of the United States of America. 102 (24): 8740–8745. Bibcode:2005PNAS..102.8740O. doi:10.1073/pnas.0503274102. ISSN 0027-8424. PMC 1143584. PMID 15939876.
- Hires, Samuel Andrew; Zhu, Yongling; Tsien, Roger Y. (2008-03-18). "Optical measurement of synaptic glutamate spillover and reuptake by linker optimized glutamate-sensitive fluorescent reporters". Proceedings of the National Academy of Sciences. 105 (11): 4411–4416. Bibcode:2008PNAS..105.4411H. doi:10.1073/pnas.0712008105. ISSN 0027-8424. PMC 2393813. PMID 18332427.
- Helassa, Nordine; Dürst, Céline D.; Coates, Catherine; Kerruth, Silke; Arif, Urwa; Schulze, Christian; Wiegert, J. Simon; Geeves, Michael; Oertner, Thomas G.; Török, Katalin (2018-05-22). "Ultrafast glutamate sensors resolve high-frequency release at Schaffer collateral synapses". Proceedings of the National Academy of Sciences. 115 (21): 5594–5599. doi:10.1073/pnas.1720648115. PMC 6003469. PMID 29735711.
- Marvin, Jonathan S.; Scholl, Benjamin; Wilson, Daniel E.; Podgorski, Kaspar; Kazemipour, Abbas; Müller, Johannes Alexander; Schoch, Susanne; Quiroz, Francisco José Urra; Rebola, Nelson; Bao, Huan; Little, Justin P. (November 2018). "Stability, affinity, and chromatic variants of the glutamate sensor iGluSnFR". Nature Methods. 15 (11): 936–939. doi:10.1038/s41592-018-0171-3. ISSN 1548-7105. PMC 6394230. PMID 30377363.
| Optogenetics | |
|---|---|
| Optogenetic actuators | |
| Optogenetic sensors | |
| Related techniques | |