| This article relies excessively on references to primary sources. Please improve this article by adding secondary or tertiary sources. Find sources: "Termite-flg RNA motif" – news · newspapers · books · scholar · JSTOR (November 2021) (Learn how and when to remove this message) |
| Termite-flg RNA | |
|---|---|
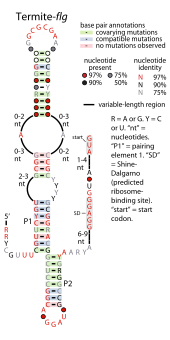 Consensus secondary structure of Termite-flg RNAs Consensus secondary structure of Termite-flg RNAs | |
| Identifiers | |
| Symbol | Termite-flg |
| Alt. Symbols | tg-flg |
| Rfam | RF01729 |
| Other data | |
| RNA type | Cis-regulatory element |
| Domain(s) | Termite gut metagenome |
| PDB structures | PDBe |
The Termite-flg RNA motif (also called tg-flg) is a conserved RNA structure identified by bioinformatics. Genomic sequences corresponding to Termite-flg RNAs have been identified only in uncultivated bacteria present in the termite hindgut. As of 2010 it has not been identified in the DNA of any cultivated species, and is thus an example of RNAs present in environmental samples.
Termite-flg RNAs are consistently located in what is presumed to be the 5' untranslated regions of genes that encode proteins whose functions relate to flagella. The RNAs are hypothesized to regulate these genes in an unknown mechanism.
References
- Weinberg Z, Perreault J, Meyer MM, Breaker RR (December 2009). "Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis". Nature. 462 (7273): 656–659. Bibcode:2009Natur.462..656W. doi:10.1038/nature08586. PMC 4140389. PMID 19956260.
- Weinberg Z, Wang JX, Bogue J, et al. (March 2010). "Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea and their metagenomes". Genome Biol. 11 (3): R31. doi:10.1186/gb-2010-11-3-r31. PMC 2864571. PMID 20230605.
External links
This molecular or cell biology article is a stub. You can help Misplaced Pages by expanding it. |