| Revision as of 08:11, 2 December 2023 editHunan201p (talk | contribs)Extended confirmed users20,357 edits The pre-print mentioned here has been published, and the published document says nothing to this effect. Removed massive synthesis of studies.← Previous edit | Latest revision as of 12:42, 9 January 2025 edit undoAnomieBOT (talk | contribs)Bots6,574,015 editsm Dating maintenance tags: {{Citation needed}} | ||
| (254 intermediate revisions by 56 users not shown) | |||
| Line 1: | Line 1: | ||
| {{short description|Genetic history of East Asian peoples}} | {{short description|Genetic history of East Asian peoples}} | ||
| {{Use dmy dates|date= |
{{Use dmy dates|date=January 2025}} | ||
| This article |
This article summarizes the genetic makeup and population history of ] and their connection to genetically related populations such as ] and ]ns, as well as ], and partly, ], ], and ]. They are collectively referred to as "]" in population genomics.<ref name=":1">{{Cite journal |last1=Bennett |first1=Andrew E. |last2=Liu |first2=Yichen |last3=Fu |first3=Qiaomei |date=4 December 2024 |title=Reconstructing the Human Population History of East Asia through Ancient Genomics |url=https://www.cambridge.org/core/elements/reconstructing-the-human-population-history-of-east-asia-through-ancient-genomics/0524D629660B5E43FC7094C043D54C6A |journal=Elements in Ancient East Asia |language=en |doi=10.1017/9781009246675|doi-access=free |isbn=978-1-009-24667-5 }}</ref> | ||
| == Overview == | == Overview == | ||
| <gallery mode="nolines" widths="275"> | |||
| ] | |||
| File:Phylogenetic structure of Eastern Eurasians.png|Phylogenetic position of East Asian lineages among other Eastern Eurasians. | |||
| ] | |||
| File:Schematic of Populations in Eurasia from 45 to 10 kaBP.png|Schematic of Populations in Eurasia from 45 to 10 kaBP. | |||
| ] | |||
| File:Schematic depicting major ancestries in Asia.png|Highlighted regions show where ancient individuals associated with the labeled ancestry have been sampled. | |||
| Population genomic studies have studied the origin and formation of modern East Asians. Ancestors of East Asians (]) split from other human populations possibly as early as 70,000 to 50,000 years ago. Possible routes into ] include a northern route model from ], beginning north of the ], and a ] model, beginning south of the Himalayas and moving through ].<ref name="Computer simulation of human leukoc"/><ref>{{cite journal | vauthors = Vallini L, Marciani G, Aneli S, Bortolini E, Benazzi S, Pievani T, Pagani L | title = Genetics and Material Culture Support Repeated Expansions into Paleolithic Eurasia from a Population Hub Out of Africa | journal = Genome Biology and Evolution | volume = 14 | issue = 4 | date = April 2022 | pmid = 35445261 | pmc = 9021735 | doi = 10.1093/gbe/evac045 }}</ref> | |||
| </gallery>] | |||
| Population genomic research has studied the origin and formation of modern East Asians. The ancestors of East Asians (]) split from other human populations possibly as early as 70,000 to 50,000 years ago. Possible routes into ] include a northern route model from ], beginning north of the ], and a ] model, beginning south of the Himalayas and moving through ].<ref>{{cite journal | vauthors = Di D, Sanchez-Mazas A, Currat M | title = Computer simulation of human leukocyte antigen genes supports two main routes of colonization by human populations in East Asia | journal = BMC Evolutionary Biology | volume = 15 | issue = 1 | pages = 240 | date = November 2015 | pmid = 26530905 | pmc = 4632674 | doi = 10.1186/s12862-015-0512-0 | bibcode = 2015BMCEE..15..240D | doi-access = free}}</ref><ref>{{cite journal | vauthors = Vallini L, Marciani G, Aneli S, Bortolini E, Benazzi S, Pievani T, Pagani L | title = Genetics and Material Culture Support Repeated Expansions into Paleolithic Eurasia from a Population Hub Out of Africa | journal = Genome Biology and Evolution | volume = 14 | issue = 4 | date = April 2022 | pmid = 35445261 | pmc = 9021735 | doi = 10.1093/gbe/evac045}}</ref><ref name=":1" /> Seguin-Orlando et al. (2014) stated that East Asians diverged from ], which occurred at least 36,200 years ago, during the ].<ref>{{Cite journal |last1=Seguin-Orlando |first1=Andaine |last2=Korneliussen |first2=Thorfinn S. |last3=Sikora |first3=Martin |last4=Malaspinas |first4=Anna-Sapfo |display-authors=3 |date=2014 |title=Genomic structure in Europeans dating back at least 36,200 years |url=https://www.science.org/doi/abs/10.1126/science.aaa0114 |journal=Science |volume=346 |issue=6213 |pages=1113–1118 |doi=10.1126/science.aaa0114 |pmid=25378462 |bibcode=2014Sci...346.1113S |via=Science.org}}</ref> Vallini et al. 2024 noted that this divergence most likely occurred on the ] 48,000 years ago.<ref>{{Cite journal |last1=Vallini |first1=Leonardo |last2=Zampieri |first2=Carlo |last3=Shoaee |first3=Mohamed Javad |last4=Bortolini |first4=Eugenio |last5=Marciani |first5=Giulia |last6=Aneli |first6=Serena |last7=Pievani |first7=Telmo |last8=Benazzi |first8=Stefano |last9=Barausse |first9=Alberto |last10=Mezzavilla |first10=Massimo |last11=Petraglia |first11=Michael D. |last12=Pagani |first12=Luca |date=25 March 2024 |title=The Persian plateau served as hub for Homo sapiens after the main out of Africa dispersal |journal=Nature Communications |language=en |volume=15 |issue=1 |pages=1882 |bibcode=2024NatCo..15.1882V |doi=10.1038/s41467-024-46161-7 |issn=2041-1723 |pmc=10963722 |pmid=38528002}}</ref> | |||
| The southern route model for East Asians has been corroborated in multiple recent studies, showing that most of the ancestry of Eastern Asians arrived from the southern route in to Southeast Asia at a very early period, starting perhaps as early as 70,000 years ago, and dispersed northward across Eastern Asia.<ref>{{Cite journal |vauthors=Osada N, Kawai Y |date=2021 |title=Exploring models of human migration to the Japanese archipelago using genome-wide genetic data |url=https://www.jstage.jst.go.jp/article/ase/129/1/129_201215/_html/-char/en |journal=Anthropological Science |volume=129 |issue=1 |pages=45–58 |doi=10.1537/ase.201215 |quote=Via the southern route, ancestors of current Asian populations reached Southeast Asia and a part of Oceania around 70000–50000 years ago, probably through a coastal dispersal route (Bae et al., 2017). The oldest samples providing the genetic evidence of the northern migration route come from a high-coverage genome sequence of individuals excavated from the Yana RHS site in northeastern Siberia (Figure 2), which is about 31600 years old (Sikora et al., 2019). A wide range of artifacts, including bone crafts of wooly rhinoceros and mammoths, were excavated at the site (Pitulko et al., 2004). The analysis of genome sequences showed that the samples were deeply diverged from most present-day East Asians and more closely related to present-day Europeans, suggesting that the population reached the area through a route different from the southern route. A 24000-year-old individual excavated near Lake Baikal (Figure 2), also known as the Mal'ta boy, and 17000-year-old individuals from the Afontova Gora II site (Afontova Gora 2 and 3) showed similar genetic features to the Yana individuals (Raghavan et al., 2014; Fu et al., 2016; Sikora et al., 2019). Interestingly, genetic data suggested that Yana individuals received a large amount of gene flow from the East Asian lineage (Sikora et al., 2019; Yang et al., 2020). |s2cid=234247309|doi-access=free}}</ref><ref name="Ancient Jomon genome sequence analy" /><ref name="auto4" /><ref>{{cite journal | vauthors = Bae CJ, Douka K, Petraglia MD | title = On the origin of modern humans: Asian perspectives | journal = Science | volume = 358 | issue = 6368 | pages = eaai9067 | date = December 2017 | pmid = 29217544 | doi = 10.1126/science.aai9067 | s2cid = 4436271 | doi-access = free }}</ref><ref>{{cite journal | vauthors = Aoki K, Takahata N, Oota H, Wakano JY, Feldman MW | title = Infectious diseases may have arrested the southward advance of microblades in Upper Palaeolithic East Asia | journal = Proceedings. Biological Sciences | volume = 290 | issue = 2005 | pages = 20231262 | date = August 2023 | pmid = 37644833 | pmc = 10465978 | doi = 10.1098/rspb.2023.1262 | quote = A single major migration of modern humans into the continents of Asia and Sahul was strongly supported by earlier studies using mitochondrial DNA, the non-recombining portion of Y chromosomes, and autosomal SNP data . Ancestral Ancient South Indians with no West Eurasian relatedness, East Asians, Onge (Andamanese hunter–gatherers) and Papuans all derive in a short evolutionary time from the eastward dispersal of an out-of-Africa population , although Europeans and East Asians are suggested to share more recent common ancestors than with Papuans . The HUGO (Human Genome Organization) Pan-Asian SNP consortium investigated haplotype diversity within present-day Asian populations and found a strong correlation with latitude, with diversity decreasing from south to north. The correlation continues to hold when only mainland Southeast Asian and East Asian populations are considered, and is perhaps attributable to a serial founder effect . These observations are consistent with the view that soon after the single eastward migration of modern humans, East Asians diverged in southern East Asia and dispersed northward across the continent. }}</ref> However, genetic evidence also supports more recent migrations to East Asia from ] along the northern route, as shown by the presence of haplogroups ] and ], as well as ] ancestry.<ref>{{cite book | vauthors = Xu D, Li H |title=Languages and Genes in Northwestern China and Adjacent Regions |date=2017 |publisher=Springer |isbn=978-981-10-4169-3 |page=27 |url=https://books.google.com/books?id=YQPNDgAAQBAJ&pg=PA27 |language=en}} "In the study of Zhong et al. haplogroups O-M175, C-M130, D-M174 and N-M231 still suggests the substantial contribution of the southern route. However, the Central Asia and West Eurasia related haplogroups, such as haplogroups R-M207 and Q-M242, occur primarily in northwestern East Asia and their frequencies gradually decrease from west to east. In addition, the Y-STR diversities of haplogroups R-M207 and Q-M242 also indicate the existence of northern route migration about 18,000 years ago from Central Asia to North Asia, and recent population admixture along the Silk Road since about 3000 years ago (Piazza 1998)."</ref><ref>{{cite journal | vauthors = Zhang X, Ji X, Li C, Yang T, Huang J, Zhao Y, Wu Y, Ma S, Pang Y, Huang Y, He Y, Su B | display-authors = 6 | title = A Late Pleistocene human genome from Southwest China | journal = Current Biology | volume = 32 | issue = 14 | pages = 3095–3109.e5 | date = July 2022 | pmid = 35839766 | doi = 10.1016/j.cub.2022.06.016 | s2cid = 250502011 | doi-access = free }} "In addition to the earliest southern settlement of AMHs in East Asia, ancient migration (40–18 kya) into East Asia via the “Northern Route” from West Eurasia was previously proposed. The “Northern Route” hypothesis would also explain where the subtle shared ancient north Eurasian (ANE) ancestry came from that is then also shared with Native Americans."</ref> | |||
| Phylogenetic data suggests that an early ] wave (>45kya) "ascribed to a population movement with uniform genetic features and material culture" (]) used a ] through ], where they subsequently diverged rapidly, and gave rise to ]s (Oceanians), the Ancient Ancestral South Indians (AASI), as well as ] and East/Southeast Asians,<ref name="Yang32" /> although ] may have also received some geneflow from an earlier group (xOoA),<ref>Genetics and material culture support repeated expansions into Paleolithic Eurasia from a population hub out of Africa, Vallini et al. 2022 (4 April 2022) Quote: "''Taken together with a lower bound of the final settlement of Sahul at 37 ka (the date of the deepest population splits estimated by Malaspinas et al. 2016), it is reasonable to describe Papuans as either an almost even mixture between East Asians and a lineage basal to West and East Asians occurred sometimes between 45 and 38 ka, or as a sister lineage of East Asians with or without a minor basal OoA or xOoA contribution. We here chose to parsimoniously describe Papuans as a simple sister group of Tianyuan, cautioning that this may be just one out of six equifinal possibilities.''"</ref> around 2%,<ref>{{cite web |title=Almost all living people outside of Africa trace back to a single migration more than 50,000 years ago |url=https://www.science.org/content/article/almost-all-living-people-outside-africa-trace-back-single-migration-more-50000-years |access-date=19 August 2022 |website=science.org |language=en}}</ref> next to additional archaic admixture in the ] region.]The southern route model for East Asians has been corroborated in multiple recent studies, showing that most of the ancestry of Eastern Asians arrived from the southern route in to Southeast Asia at a very early period, starting perhaps as early as 70,000 years ago, and dispersed northward across Eastern Asia.<ref name=":2" /><ref name="Ancient Jomon genome sequence analy" /><ref name="auto4" /><ref>{{cite journal | vauthors = Bae CJ, Douka K, Petraglia MD | title = On the origin of modern humans: Asian perspectives | journal = Science | volume = 358 | issue = 6368 | pages = eaai9067 | date = December 2017 | pmid = 29217544 | doi = 10.1126/science.aai9067 | s2cid = 4436271 | doi-access = free}}</ref><ref>{{cite journal | vauthors = Aoki K, Takahata N, Oota H, Wakano JY, Feldman MW | title = Infectious diseases may have arrested the southward advance of microblades in Upper Palaeolithic East Asia | journal = Proceedings. Biological Sciences | volume = 290 | issue = 2005 | pages = 20231262 | date = August 2023 | pmid = 37644833 | pmc = 10465978 | doi = 10.1098/rspb.2023.1262 | quote = A single major migration of modern humans into the continents of Asia and Sahul was strongly supported by earlier studies using mitochondrial DNA, the non-recombining portion of Y chromosomes, and autosomal SNP data . Ancestral Ancient South Indians with no West Eurasian relatedness, East Asians, Onge (Andamanese hunter–gatherers) and Papuans all derive in a short evolutionary time from the eastward dispersal of an out-of-Africa population , although Europeans and East Asians are suggested to share more recent common ancestors than with Papuans . The HUGO (Human Genome Organization) Pan-Asian SNP consortium investigated haplotype diversity within present-day Asian populations and found a strong correlation with latitude, with diversity decreasing from south to north. The correlation continues to hold when only mainland Southeast Asian and East Asian populations are considered, and is perhaps attributable to a serial founder effect . These observations are consistent with the view that soon after the single eastward migration of modern humans, East Asians diverged in southern East Asia and dispersed northward across the continent.}}</ref><ref>{{Cite journal |last1=Demeter |first1=Fabrice |last2=Shackelford |first2=Laura L. |last3=Bacon |first3=Anne-Marie |last4=Duringer |first4=Philippe |last5=Westaway |first5=Kira |last6=Sayavongkhamdy |first6=Thongsa |last7=Braga |first7=José |last8=Sichanthongtip |first8=Phonephanh |last9=Khamdalavong |first9=Phimmasaeng |last10=Ponche |first10=Jean-Luc |last11=Wang |first11=Hong |last12=Lundstrom |first12=Craig |last13=Patole-Edoumba |first13=Elise |last14=Karpoff |first14=Anne-Marie |date=4 September 2012 |title=Anatomically modern human in Southeast Asia (Laos) by 46 ka |journal=Proceedings of the National Academy of Sciences |language=en |volume=109 |issue=36 |pages=14375–14380 |doi=10.1073/pnas.1208104109 |doi-access=free |issn=0027-8424 |pmc=3437904 |pmid=22908291 |bibcode=2012PNAS..10914375D |quote=Inferences from nuclear (51), Y chromosome (52), and mitochondrial genome (53) data support an early migration of modern humans out of Africa and into Southeast Asia using a southern route by at least 60 ka. Patterns of genetic variation in recent human populations (11, 54, 55) recognize Southeast Asia as an important source for the peopling of East Asia and Australasia via a rapid, early settlement.}}</ref><ref name=":1" /> However, genetic evidence also supports more recent migrations to East Asia from ] and ] along the northern route, as shown by the presence of haplogroups ] and ], as well as ] ancestry.<ref>{{cite book | vauthors = Xu D, Li H |title=Languages and Genes in Northwestern China and Adjacent Regions |date=2017 |publisher=Springer |isbn=978-981-10-4169-3 |page=27 |url=https://books.google.com/books?id=YQPNDgAAQBAJ&pg=PA27 |language=en}} "In the study of Zhong et al. haplogroups O-M175, C-M130, D-M174 and N-M231 still suggests the substantial contribution of the southern route. However, the Central Asia and West Eurasia related haplogroups, such as haplogroups R-M207 and Q-M242, occur primarily in northwestern East Asia and their frequencies gradually decrease from west to east. In addition, the Y-STR diversities of haplogroups R-M207 and Q-M242 also indicate the existence of northern route migration about 18,000 years ago from Central Asia to North Asia, and recent population admixture along the Silk Road since about 3000 years ago (Piazza 1998)."</ref><ref>{{cite journal | vauthors = Zhang X, Ji X, Li C, Yang T, Huang J, Zhao Y, Wu Y, Ma S, Pang Y, Huang Y, He Y, Su B | display-authors = 6 | title = A Late Pleistocene human genome from Southwest China | journal = Current Biology | volume = 32 | issue = 14 | pages = 3095–3109.e5 | date = July 2022 | pmid = 35839766 | doi = 10.1016/j.cub.2022.06.016 | s2cid = 250502011 | doi-access = free | bibcode = 2022CBio...32E3095Z}} "In addition to the earliest southern settlement of AMHs in East Asia, ancient migration (40–18 kya) into East Asia via the “Northern Route” from West Eurasia was previously proposed. The “Northern Route” hypothesis would also explain where the subtle shared ancient north Eurasian (ANE) ancestry came from that is then also shared with Native Americans."</ref> | |||
| The southern migration wave likely diversified after settling within East Asia, while the northern wave, which probably arrived from the Eurasian steppe, mixed with the southern wave, probably in Siberia.<ref name="Sato evab192" /> | |||
| ] | |||
| A review paper by Melinda A. Yang (in 2022) summarized and concluded that a distinctive "Basal-East Asian population" referred to as ''East- and Southeast Asian lineage'' (ESEA); which is ancestral to modern ], ]ns, ], and ], originated in ] at c. 50,000 BCE, and expanded through multiple migration waves southwards and northwards, respectively. The ESEA lineage is also ancestral to the "basal" ] of Southeast Asia and the c. 40,000-year-old ] lineage found in ], which can already be differentiated from the deeply related Ancestral Ancient South Indians (AASI) and ]n (AA) lineages.<ref name="Yang">{{Cite journal | vauthors = Yang MA |date=6 January 2022 |title=A genetic history of migration, diversification, and admixture in Asia |url=http://www.pivotscipub.com/hpgg/2/1/0001 |journal=Human Population Genetics and Genomics |language=en |volume=2 |issue=1 |pages=1–32 |doi=10.47248/hpgg2202010001 |issn=2770-5005 |quote=...In contrast, mainland East and Southeast Asians and other Pacific islanders (e.g., Austronesian speakers) are closely related to each other and here denoted as belonging to an East and Southeast Asian (ESEA) lineage (Box 2). …the ESEA lineage differentiated into at least three distinct ancestries: Tianyuan ancestry which can be found 40,000–33,000 years ago in northern East Asia, ancestry found today across present-day populations of East Asia, Southeast Asia, and Siberia, but whose origins are unknown, and Hòabìnhian ancestry found 8,000–4,000 years ago in Southeast Asia, but whose origins in the Upper Paleolithic are unknown.|doi-access=free}}</ref> There are currently eight detected, closely related, sub-ancestries in the ESEA lineage: | |||
| The southern migration wave likely diversified after settling within East Asia, while the northern wave, which probably arrived from the Eurasian steppe, mixed with the southern wave, probably in Siberia.<ref name="Sato evab192" /> | |||
| * ] ancestry – Associated with populations in the ] region, ], and ], as well as parts of ]. | |||
| * ] ancestry – Associated with ancient samples in the ] region of ], and modern ]. | |||
| A review paper by Melinda A. Yang (in 2022) described the ''East- and Southeast Asian lineage'' (ESEA); which is ancestral to modern ], ]ns, ], and ], originated in ] at c. 50,000 BCE, and expanded through multiple migration waves southwards and northwards, respectively. The ESEA lineage is also ancestral to the "basal Asian" ] of Southeast Asia and the c. 40,000-year-old ] lineage found in ], which can already be differentiated from the deeply related Ancestral Ancient South Indians (AASI) and ]n (AA) lineages.<ref name="Yang32" /> There are currently eight detected, closely related, sub-ancestries in the ESEA lineage: | |||
| * ] ancestry – Associated with a 10,500-year-old individual from ], ]. | |||
| * ] (ANA) – Associated with populations in the ] region, ], and ], as well as parts of ]. | |||
| * ] – Associated with ancient samples in the ] region of ], and modern ]. | |||
| * ] ancestry – Associated with a 10,500-year-old individual from ], ]. This ancestry was not observed in either historical samples from Guangxi or contemporary East and Southeast Asians, suggesting that the lineage is extinct in the modern day. | |||
| * ] ancestry – Ancestry associated with 8,000–3,000-year-old individuals in the ]. | * ] ancestry – Ancestry associated with 8,000–3,000-year-old individuals in the ]. | ||
| * ] ancestry – Ancestry on the ESEA lineage associated with 8,000–4,000-year-old hunter-gatherers in ] and ]. | * ] – Ancestry on the ESEA lineage associated with 8,000–4,000-year-old hunter-gatherers in ] and ]. | ||
| * ] ancestry – Ancestry on the ESEA lineage associated with an ] individual dating to 40,000 years ago in ]. | * ] ancestry – Ancestry on the ESEA lineage associated with an ] individual dating to 40,000 years ago in ]. | ||
| * ]an ancestry – Associated with 3,000-year-old individuals in the ] region of the ]. | * ]an ancestry – Associated with 3,000-year-old individuals in the ] region of the ]. | ||
| * ] ancestry – Associated with populations in the ] region and common among ]. | * ] ancestry – Associated with populations in the ] region and common among ]. | ||
| ]The genetic makeup of East Asians is primarily characterized by "Yellow River" (East Asian) ancestry which formed from a major ] (ANEA) component and a minor ] (ASEA) one. The two lineages diverged from each other at least 19,000 years ago, after the divergence of the ], ] (Longlin), ] and ].<ref name="sciencedirect.com" /><ref name="Yang32" /> Contemporary East Asians (notably Sino-Tibetan speakers) mostly have Yellow River ancestry, which is associated with millet and rice cultivation. "East Asian Highlanders" (Tibetans) carry both Tibetan ancestry and Yellow River ancestry. Japanese people were found to have a tripartite origin; consisting of Jōmon ancestry, Amur ancestry, and Yellow River ancestry.<ref>{{cite bioRxiv |biorxiv=10.1101/2020.12.07.414037 |title=Comprehensive analysis of Japanese archipelago population history by detecting ancestry-marker polymorphisms without using ancient DNA data |date=8 March 2021 |vauthors=Watanabe Y, Ohashi J}}</ref><ref>{{cite journal |vauthors=de Boer E |date=2020 |title=Japan considered from the hypothesis of farmer/language spread |journal=Evolutionary Human Sciences |volume=2 |pages=e13 |doi=10.1017/ehs.2020.7 |pmc=10427481 |pmid=37588377 |s2cid=218926428 |doi-access=free}}</ref> East Asians carry a variation of the ''MFSD12'' gene, which is responsible for lighter skin colour.<ref>{{Cite press release |title=Genetic study provides novel insights into the evolution of skin colour |date=21 January 2019 |publisher=University College London |url=https://www.ucl.ac.uk/news/2019/jan/genetic-study-provides-novel-insights-evolution-skin-colour |language=en |last=Downes |first=Natasha |access-date=4 December 2021 |work=UCL News}}</ref> Huang et al. (2021) found evidence for light skin being ] among the ancestral populations of West Eurasians and East Eurasians, prior to their divergence.<ref>{{cite journal |last1=Huang |first1=Xin |date=2021 |title=Dissecting dynamics and differences of selective pressures in the evolution of human pigmentation |journal=Biology Open |volume=10 |issue=2 |doi=10.1242/bio.056523 |pmc=7888712 |pmid=33495209}}</ref> | |||
| Northeast Asians such as ], ], and ] peoples derive most of their ancestry from the "Amur" (]) subgroup of the ]s, which expanded massively with ] cultivation and ]. Tungusic peoples display the highest genetic affinity to Ancient Northeast Asians, represented by c. 7,000 and 13,000 year old specimens, whereas Turkic peoples have significant West Eurasian admixture.<ref>{{Cite journal |last1=He |first1=Guang-Lin |last2=Wang |first2=Meng-Ge |last3=Zou |first3=Xing |last4=Yeh |first4=Hui-Yuan |last5=Liu |first5=Chang-Hui |last6=Liu |first6=Chao |last7=Chen |first7=Gang |last8=Wang |first8=Chuan-Chao |date= 2023|title=Extensive ethnolinguistic diversity at the crossroads of North China and South Siberia reflects multiple sources of genetic diversity |url=https://onlinelibrary.wiley.com/doi/10.1111/jse.12827 |journal=Journal of Systematics and Evolution |language=en |volume=61 |issue=1 |pages=230–250 |doi=10.1111/jse.12827 |issn=1674-4918}}</ref><ref>{{cite journal |display-authors=6 |vauthors=Mao X, Zhang H, Qiao S, Liu Y, Chang F, Xie P, Zhang M, Wang T, Li M, Cao P, Yang R, Liu F, Dai Q, Feng X, Ping W, Lei C, Olsen JW, Bennett EA, Fu Q |date=June 2021 |title=The deep population history of northern East Asia from the Late Pleistocene to the Holocene |journal=Cell |language=English |volume=184 |issue=12 |pages=3256–3266.e13 |doi=10.1016/j.cell.2021.04.040 |pmid=34048699 |s2cid=235226413 |doi-access=free}}</ref> | |||
| Modern Northeast Asians consist mostly of the "Amur ancestry" which expanded massively with ] cultivation. Modern Southeast Asians (specifically Austronesians) consist mainly of the "Fujian ancestry" component, which is associated with the spread of ]. Contemporary East Asians (most notably Sino-Tibetan speakers) consist mostly of Yellow River ancestry, associated with both millet and rice cultivation. "East Asian Highlanders" (Tibetans) consist of both the Ancient Tibetan ancestry and Yellow River ancestry. Japanese people were found to have a tripartite origin; consisting of Jōmon ancestry, Amur ancestry, and Yellow River ancestry. ] formed from Ancient North Eurasians and from an early East Asian branch, giving rise to "Ancestral Beringians", which gave rise to both "Paleosiberians" and contemporary ]. Isolated hunter-gatherer in Southeast Asia, specifically Malaysia and Thailand, such as the ], derive most of their ancestry from the Hoabinhian lineage.<ref name="Ancient Jomon genome sequence analy" /><ref>{{cite journal |display-authors=6 |vauthors=Wong EH, Khrunin A, Nichols L, Pushkarev D, Khokhrin D, Verbenko D, Evgrafov O, Knowles J, Novembre J, Limborska S, Valouev A |date=January 2017 |title=Reconstructing genetic history of Siberian and Northeastern European populations |journal=Genome Research |volume=27 |issue=1 |pages=1–14 |doi=10.1101/gr.202945.115 |pmc=5204334 |pmid=27965293}}</ref><ref name="auto1" /><ref>{{cite journal |display-authors=6 |vauthors=Mao X, Zhang H, Qiao S, Liu Y, Chang F, Xie P, Zhang M, Wang T, Li M, Cao P, Yang R, Liu F, Dai Q, Feng X, Ping W, Lei C, Olsen JW, Bennett EA, Fu Q |date=June 2021 |title=The deep population history of northern East Asia from the Late Pleistocene to the Holocene |journal=Cell |language=English |volume=184 |issue=12 |pages=3256–3266.e13 |doi=10.1016/j.cell.2021.04.040 |pmid=34048699 |s2cid=235226413|doi-access=free}}</ref><ref name="auto5">{{cite journal |vauthors=Tagore D, Aghakhanian F, Naidu R, Phipps ME, Basu A |date=March 2021 |title=Insights into the demographic history of Asia from common ancestry and admixture in the genomic landscape of present-day Austroasiatic speakers |journal=BMC Biology |volume=19 |issue=1 |pages=61 |doi=10.1186/s12915-021-00981-x |pmc=8008685 |pmid=33781248 |doi-access=free }}</ref><ref>{{cite web |date=3 January 2018 |title=Direct genetic evidence of founding population reveals story of first Native Americans |url=https://www.cam.ac.uk/research/news/direct-genetic-evidence-of-founding-population-reveals-story-of-first-native-americans |access-date=9 June 2021 |website=University of Cambridge |language=en}}</ref><ref>{{cite journal |vauthors=Liu D, Duong NT, Ton ND, Van Phong N, Pakendorf B, Van Hai N, Stoneking M |date=September 2020 |title=Extensive Ethnolinguistic Diversity in Vietnam Reflects Multiple Sources of Genetic Diversity |journal=Molecular Biology and Evolution |volume=37 |issue=9 |pages=2503–2519 |doi=10.1093/molbev/msaa099 |pmc=7475039 |pmid=32344428}}</ref><ref>{{cite bioRxiv |biorxiv=10.1101/2020.12.07.414037 |title=Comprehensive analysis of Japanese archipelago population history by detecting ancestry-marker polymorphisms without using ancient DNA data |date=8 March 2021 |vauthors=Watanabe Y, Ohashi J}}</ref><ref>{{cite journal |vauthors=de Boer E |date=2020 |title=Japan considered from the hypothesis of farmer/language spread |journal=Evolutionary Human Sciences |volume=2 |pages=e13 |doi=10.1017/ehs.2020.7 |pmid=37588377 |pmc=10427481 |s2cid=218926428|doi-access=free}}</ref> | |||
| East Asian populations exhibit some European-related admixture, originating from ] traders and interactions with Mongolians, who were well-acquainted with European-like populations. This is more common among northern Han Chinese (2.8%) than southern Han Chinese (1.7%), Japanese (2.2%), and Koreans (1.6%). However, East Asians have less European-related admixture than Northeast Asians like Mongolians (10.9%), ] (9.6%), ] (8.0%), and ] (6.8%).<ref>{{Cite journal |last1=Qin |first1=Pengfei |last2=Zhou |first2=Ying |last3=Lou |first3=Haiyi |last4=Lu |first4=Dongsheng |last5=Yang |first5=Xiong |last6=Wang |first6=Yuchen |last7=Jin |first7=Li |last8=Chung |first8=Yeun-Jun |last9=Xu |first9=Shuhua |date=2 April 2015 |title=Quantitating and Dating Recent Gene Flow between European and East Asian Populations |journal=Scientific Reports |language=en |volume=5 |issue=1 |pages=9500 |doi=10.1038/srep09500 |issn=2045-2322 |pmc=4382708 |pmid=25833680|bibcode=2015NatSR...5.9500Q }}</ref> | |||
| ] | |||
| Phylogenetic data suggests that an early initial ] or "eastern non-African" (ENA) meta-population trifurcated, and gave rise to ]s (Oceanians), the Ancient Ancestral South Indians (AASI), as well as East/Southeast Asians, although ] may have also received some geneflow from an earlier group (xOoA),<ref name="auto6">Genetics and material culture support repeated expansions into Paleolithic Eurasia from a population hub out of Africa, Vallini et al. 2022 (4 April 2022) Quote: "''Taken together with a lower bound of the final settlement of Sahul at 37 ka (the date of the deepest population splits estimated by Malaspinas et al. 2016), it is reasonable to describe Papuans as either an almost even mixture between East Asians and a lineage basal to West and East Asians occurred sometimes between 45 and 38 ka, or as a sister lineage of East Asians with or without a minor basal OoA or xOoA contribution. We here chose to parsimoniously describe Papuans as a simple sister group of Tianyuan, cautioning that this may be just one out of six equifinal possibilities.''"</ref> around 2%,<ref name="auto7">{{cite web |title=Almost all living people outside of Africa trace back to a single migration more than 50,000 years ago |url=https://www.science.org/content/article/almost-all-living-people-outside-africa-trace-back-single-migration-more-50000-years |access-date=19 August 2022 |website=science.org |language=en}}</ref> next to additional archaic admixture in the ] region. | |||
| Austronesians mainly carry "Fujian" (]) ancestry, which is associated with the spread of ]. Isolated hunter-gatherers in Southeast Asia, specifically in Malaysia and Thailand, such as the ], derive most of their ancestry from the Hoabinhian lineage.<ref name="auto5" /><ref>{{cite web |date=3 January 2018 |title=Direct genetic evidence of founding population reveals story of first Native Americans |url=https://www.cam.ac.uk/research/news/direct-genetic-evidence-of-founding-population-reveals-story-of-first-native-americans |access-date=9 June 2021 |website=University of Cambridge |language=en}}</ref><ref>{{cite journal |vauthors=Liu D, Duong NT, Ton ND, Van Phong N, Pakendorf B, Van Hai N, Stoneking M |date=September 2020 |title=Extensive Ethnolinguistic Diversity in Vietnam Reflects Multiple Sources of Genetic Diversity |journal=Molecular Biology and Evolution |volume=37 |issue=9 |pages=2503–2519 |doi=10.1093/molbev/msaa099 |pmc=7475039 |pmid=32344428}}</ref> The emergence of the ] went along with a population shift caused by migrations from southern China. Neolithic Mainland Southeast Asian samples predominantly have Ancient Southern East Asian ancestry with Hoabinhian-related admixture. In modern populations, this admixture of Ancient Southern East Asian and Hoabinhian ancestry is most strongly associated with ] speakers.<ref>{{Citation |last1=Liu |first1=Dang |title=Extensive ethnolinguistic diversity in Vietnam reflects multiple sources of genetic diversity |date=28 November 2019 |url=https://www.biorxiv.org/content/10.1101/857367v1 |access-date=14 November 2024 |language=en |doi=10.1101/857367 |last2=Duong |first2=Nguyen Thuy |last3=Ton |first3=Nguyen Dang |last4=Phong |first4=Nguyen Van |last5=Pakendorf |first5=Brigitte |last6=Hai |first6=Nong Van |last7=Stoneking |first7=Mark|hdl=21.11116/0000-0006-4AD8-4 |hdl-access=free }}</ref> | |||
| == Xiongnu people (ancient) == | |||
| {{Main|Xiongnu}} | |||
| The ], possibly a ], ], ] or multi-ethnic people, were a ]<ref>{{cite web |title=Xiongnu People |url=http://global.britannica.com/topic/Xiongnu |url-status=dead |archive-url=https://web.archive.org/web/20200311191625/https://global.britannica.com/topic/Xiongnu |archive-date=11 March 2020 |access-date=25 July 2015 |website=britannica.com |publisher=Encyclopædia Britannica}}</ref> of ] peoples who, according to ancient ], inhabited the eastern ] from the 3rd century BC to the late 1st century AD. ] report that ], the supreme leader after 209 BC, founded the Xiongnu Empire.<ref>di Cosmo 2004: 186</ref> | |||
| An early branch of Ancient Northern East Asians, together with ], gave rise to the ], who in turn gave rise to both "modern Paleo-Siberians" (such as ], ], and ] speakers) and contemporary ]. Paleo-Siberian ancestry was once widespread across ], but it was largely replaced by Neo-Siberian ancestry due to a major population turnover from the south, possibly involving ] and ] speakers. This was later followed by another expansion from the south in relatively recent times, associated with Amur River ancestry involving Tungusic, Mongolic, and Turkic speakers. | |||
| According to data compiled from several genetic studies, Xiongnu samples were found to have approximately 58% East Eurasian ancestry, represented by a bronze-age population from ], Mongolia. The rest of the Xiongnu's ancestry (~40%) was related to West Eurasians, represented by the ] ] population of Central Asia, and the ] culture of the Western steppe.<ref>{{cite journal | vauthors = Savelyev A, Jeong C | title = Early nomads of the Eastern Steppe and their tentative connections in the West | journal = Evolutionary Human Sciences | volume = 2 | date = May 2020 | pmid = 35663512 | pmc = 7612788 | doi = 10.1017/ehs.2020.18 | s2cid = 218935871 | hdl = 21.11116/0000-0007-772B-4 }}</ref><ref>{{harvnb|Savelyev|Jeong|2020|ps=:"Specifically, individuals from Iron Age steppe and Xiongnu have an ancestry related to present-day and ancient Iranian/Caucasus/Turan populations in addition to the ancestry components derived from the Late Bronze Age populations. We estimate that they derive between 5 and 25% of their ancestry from this new source, with 18% for Xiongnu (Table 2). We speculate that the introduction of this new western Eurasian ancestry may be linked to the Iranian elements in the Xiongnu linguistic material, while the Turkic-related component may be brought by their eastern Eurasian genetic substratum." Table 2: Sintashta_MLBA, 0.239; Khovsgol LBA, 0.582; Gonur1 BA 0.178}}</ref> | |||
| <ref>{{cite journal |display-authors=6 |vauthors=Wong EH, Khrunin A, Nichols L, Pushkarev D, Khokhrin D, Verbenko D, Evgrafov O, Knowles J, Novembre J, Limborska S, Valouev A |date=January 2017 |title=Reconstructing genetic history of Siberian and Northeastern European populations |journal=Genome Research |volume=27 |issue=1 |pages=1–14 |doi=10.1101/gr.202945.115 |pmc=5204334 |pmid=27965293}}</ref><ref>{{Citation |last1=Sikora |first1=Martin |title=The population history of northeastern Siberia since the Pleistocene |date=22 October 2018 |url=https://www.biorxiv.org/content/10.1101/448829v1 |access-date=14 November 2024 |language=en |doi=10.1101/448829 |last2=Pitulko |first2=Vladimir V. |last3=Sousa |first3=Vitor C. |last4=Allentoft |first4=Morten E. |last5=Vinner |first5=Lasse |last6=Rasmussen |first6=Simon |last7=Margaryan |first7=Ashot |last8=Damgaard |first8=Peter de Barros |last9=Castro |first9=Constanza de la Fuente|hdl=1887/3198847 |hdl-access=free }}</ref> | |||
| === Paternal lineages === | |||
| A review of the available research has shown that, as a whole, 53% of Xiongnu paternal haplogroups were East Eurasian, while 47% were West Eurasian.{{sfn|Rogers|Kaestle|2022}} | |||
| == Ancient and historical populations == | |||
| In 2012, Chinese researchers published an analysis of the paternal haplogroups of 12 elite Xiongnu male specimens from Heigouliang in ], China. Six of the specimens belonged to ], while four belonged to ]. 2 belonged to unidentified clades of Q*.<ref>{{cite report |title=Y-Chromosome Genetic Diversity of the Ancient North Chinese populations | vauthors = Li H |date=2012 |publisher=Jilin University |location=China}}</ref> In another study, a probable ] of the Xiongnu empire was assigned to ].<ref>{{cite journal | vauthors = Lee J, Miller BK, Bayarsaikhan J, Johannesson E, Ventresca Miller A, Warinner C, Jeong C | title = Genetic population structure of the Xiongnu Empire at imperial and local scales | journal = Science Advances | volume = 9 | issue = 15 | pages = eadf3904 | date = April 2023 | pmid = 37058560 | pmc = 10104459 | doi = 10.1126/sciadv.adf3904 | publisher = American Association for the Advancement of Science (AAAS) | bibcode = 2023SciA....9F3904L }}</ref><ref>{{harvnb|Lee|2023|ps=: Haplogroup information for DA39 is located in Supplementary Materials document Data S1 (abk1900_Data_S1.xlsx), row 58. | |||
| === Xiongnu people === | |||
| "Before this study, only one other individual from an elite square tomb had been analyzed in a genome-wide manner: DA39 from Tomb 1 at the imperial elite site of Gol Mod 2 in central-north Mongolia (13). This adult male, buried in one of the largest square tomb complexes excavated to date, surrounded by at least 27 satellite burials, and containing rare exotic items such as Roman glass bowls, was likely a chanyu, or ruler of the empire (73, 74)."}}</ref> | |||
| {{Main|Xiongnu}} | |||
| The ], possibly a ], ], ] or multi-ethnic people, were a ]<ref>{{cite web |title=Xiongnu People |url=http://global.britannica.com/topic/Xiongnu |url-status=dead |archive-url=https://web.archive.org/web/20200311191625/https://global.britannica.com/topic/Xiongnu |archive-date=11 March 2020 |access-date=25 July 2015 |website=britannica.com |publisher=Encyclopædia Britannica}}</ref> of ] peoples who, according to ancient ], inhabited the eastern ] from the 3rd century BC to the late 1st century AD. ] report that ], the supreme leader after 209 BC, founded the Xiongnu Empire.<ref>di Cosmo 2004: 186</ref> | |||
| === |
==== Autosomal DNA ==== | ||
| It was found that the "predominant part of the Xiongnu population is likely to have spoken Turkic". However, important cultural, technological and political elements may have been transmitted by Eastern ]-speaking Steppe nomads: "Arguably, these Iranian-speaking groups were assimilated over time by the predominant Turkic-speaking part of the Xiongnu population".<ref name="Savelyev & Jeong, 2020" /> This is reflected by the average genetic makeup of Xiongnu samples, having approximately 58% East Eurasian ancestry, represented by a Bronze Age population from ], Mongolia, which may be associated with the Turkic linguistic heritage. The rest of the Xiongnu's ancestry (~40%) was related to West Eurasians, represented by the ] ] population of Central Asia, and the ] culture of the Western steppe.<ref name="Savelyev & Jeong, 2020" /><ref>{{harvnb|Savelyev|Jeong|2020|ps=:"Specifically, individuals from Iron Age steppe and Xiongnu have an ancestry related to present-day and ancient Iranian/Caucasus/Turan populations in addition to the ancestry components derived from the Late Bronze Age populations. We estimate that they derive between 5 and 25% of their ancestry from this new source, with 18% for Xiongnu (Table 2). We speculate that the introduction of this new western Eurasian ancestry may be linked to the Iranian elements in the Xiongnu linguistic material, while the Turkic-related component may be brought by their eastern Eurasian genetic substratum." Table 2: Sintashta_MLBA, 0.239; Khovsgol LBA, 0.582; Gonur1 BA 0.178}}</ref> The Xiongnu displayed striking heterogeneity and could be differentiated into two subgroups, "Western Xiongnu" and "Eastern Xiongnu", with the former being of "hybrid" origins displaying affinity to previous ] tribes, such as represented by the ], while the later was of primarily ] (]-]) origin.<ref name=":5" /><ref name="Savelyev & Jeong, 2020" /> High status Xiongnu individuals tended to have less genetic diversity, and their ancestry was essentially derived from the Eastern Eurasian ]/].<ref>{{Cite journal |last1=Lee |first1=Juhyeon |last2=Miller |first2=Bryan K. |last3=Bayarsaikhan |first3=Jamsranjav |last4=Johannesson |first4=Erik |last5=Miller |first5=Alicia Ventresca |last6=Warinner |first6=Christina |last7=Jeong |first7=Choongwon |date=April 2023 |title=Genetic population structure of the Xiongnu Empire at imperial and local scales |journal=Science Advances |language=en |volume=9 |issue=15 |pages=eadf3904 |doi=10.1126/sciadv.adf3904 |pmid=37058560|pmc=10104459 |bibcode=2023SciA....9F3904L}}</ref> | |||
| The bulk of the genetics research indicates that, as a whole, 73% of Xiongnu maternal haplogroups were East Eurasian, while 27% were West Eurasian.<ref>{{cite journal | vauthors = Rogers LL, Kaestle FA |title=Analysis of mitochondrial DNA haplogroup frequencies in the population of the slab burial mortuary culture of Mongolia (ca. 1100–300 BCE ) |journal=American Journal of Biological Anthropology |date=2022 |volume=177 |issue=4 |pages=644–657 |doi=10.1002/ajpa.24478 |s2cid=246508594 |language=en |issn=2692-7691|doi-access=free }} " The first pattern is that the slab burial mtDNA frequencies are extremely similar to those of the aggregated Xiongnu populations and relatively similar to those of the various Bronze Age Mongolian populations, strongly supporting a population continuity hypothesis for the region over these time periods (Honeychurch, 2013)"</ref> | |||
| ==== Paternal lineages ==== | |||
| A 2003 study found that 89% of Xiongnu maternal lineages from the ] valley were of East Asian origin, while 11% were of West Eurasian origin.<ref name="Keyser-Tracqui_et_al">{{Cite journal |vauthors=Keyser-Tracqui C, Crubézy E, Ludes B |date=August 2003 |title=Nuclear and mitochondrial DNA analysis of a 2,000-year-old necropolis in the Egyin Gol Valley of Mongolia |journal=American Journal of Human Genetics |volume=73 |issue=2 |pages=247–60 |doi=10.1086/377005 |pmc=1180365 |pmid=12858290}}</ref> A 2016 study of Xiongnu from central Mongolia found a considerably higher frequency of West Eurasian maternal lineages, at 37.5%.<ref>{{cite journal | vauthors = Lee JY, Kuang S |title=A Comparative Analysis of Chinese Historical Sources and Y-DNA Studies with Regard to the Early and Medieval Turkic Peoples |journal=Inner Asia |date=2017 |volume=19 |issue=2 |pages=197–239 |doi=10.1163/22105018-12340089 |s2cid=165623743 |url=https://brill.com/view/journals/inas/19/2/article-p197_197.xml |issn=1464-8172 |doi-access=free}} "Analysis of the mitochondrial DNA, which is maternally inherited, shows that the Xiongnu remains from this Egyin Gol necropolis consist mainly of Asian lineages (89%). West Eurasian lineages makeup the rest (11%) (Keyser-Tracqui et al. (2003: 258). However, according to a more recent study of ancient human remains from central Mongolia, the Xiongnu population in central Mongolia possessed a higher frequency of western mitochondrial DNA haplotypes (37.5%) than the Xiongnu from the Egyin Gol necropolis (Rogers 2016: 78)."</ref> | |||
| A review of the available research has shown that, as a whole, 53% of Xiongnu paternal haplogroups were East Eurasian, while 47% were West Eurasian.{{sfn|Rogers|Kaestle|2022}} In 2012, Chinese researchers published an analysis of the paternal haplogroups of 12 elite Xiongnu male specimens from Heigouliang in ], China. Six of the specimens belonged to ], while four belonged to ]. 2 belonged to unidentified clades of Q*.<ref>{{cite report |title=Y-Chromosome Genetic Diversity of the Ancient North Chinese populations | vauthors = Li H |date=2012 |publisher=Jilin University |location=China}}</ref> In another study, a probable ] of the Xiongnu empire was assigned to ].<ref>{{cite journal | vauthors = Lee J, Miller BK, Bayarsaikhan J, Johannesson E, Ventresca Miller A, Warinner C, Jeong C | title = Genetic population structure of the Xiongnu Empire at imperial and local scales | journal = Science Advances | volume = 9 | issue = 15 | pages = eadf3904 | date = April 2023 | pmid = 37058560 | pmc = 10104459 | doi = 10.1126/sciadv.adf3904 | publisher = American Association for the Advancement of Science (AAAS) | bibcode = 2023SciA....9F3904L}}</ref><ref>{{harvnb|Lee|2023|ps=: Haplogroup information for DA39 is located in Supplementary Materials document Data S1 (abk1900_Data_S1.xlsx), row 58. "Before this study, only one other individual from an elite square tomb had been analyzed in a genome-wide manner: DA39 from Tomb 1 at the imperial elite site of Gol Mod 2 in central-north Mongolia (13). This adult male, buried in one of the largest square tomb complexes excavated to date, surrounded by at least 27 satellite burials, and containing rare exotic items such as Roman glass bowls, was likely a chanyu, or ruler of the empire (73, 74)."}}</ref> | |||
| ==== Maternal lineages ==== | |||
| == Xianbei people (ancient) == | |||
| The bulk of the genetics research indicates that, as a whole, 73% of Xiongnu maternal haplogroups were East Eurasian, while 27% were West Eurasian.<ref>{{cite journal | vauthors = Rogers LL, Kaestle FA |title=Analysis of mitochondrial DNA haplogroup frequencies in the population of the slab burial mortuary culture of Mongolia (ca. 1100–300 BCE ) |journal=American Journal of Biological Anthropology |date=2022 |volume=177 |issue=4 |pages=644–657 |doi=10.1002/ajpa.24478 |s2cid=246508594 |language=en |issn=2692-7691|doi-access=free}} " The first pattern is that the slab burial mtDNA frequencies are extremely similar to those of the aggregated Xiongnu populations and relatively similar to those of the various Bronze Age Mongolian populations, strongly supporting a population continuity hypothesis for the region over these time periods (Honeychurch, 2013)"</ref> A 2003 study found that 89% of Xiongnu maternal lineages from the ] valley were of East Asian origin, while 11% were of West Eurasian origin.<ref>{{Cite journal |vauthors=Keyser-Tracqui C, Crubézy E, Ludes B |date=August 2003 |title=Nuclear and mitochondrial DNA analysis of a 2,000-year-old necropolis in the Egyin Gol Valley of Mongolia |journal=American Journal of Human Genetics |volume=73 |issue=2 |pages=247–60 |doi=10.1086/377005 |pmc=1180365 |pmid=12858290}}</ref> A 2016 study of Xiongnu from central Mongolia found a considerably higher frequency of West Eurasian maternal lineages, at 37.5%.<ref>{{cite journal | vauthors = Lee JY, Kuang S |title=A Comparative Analysis of Chinese Historical Sources and Y-DNA Studies with Regard to the Early and Medieval Turkic Peoples |journal=Inner Asia |date=2017 |volume=19 |issue=2 |pages=197–239 |doi=10.1163/22105018-12340089 |s2cid=165623743 |issn=1464-8172 |doi-access=free}} "Analysis of the mitochondrial DNA, which is maternally inherited, shows that the Xiongnu remains from this Egyin Gol necropolis consist mainly of Asian lineages (89%). West Eurasian lineages makeup the rest (11%) (Keyser-Tracqui et al. (2003: 258). However, according to a more recent study of ancient human remains from central Mongolia, the Xiongnu population in central Mongolia possessed a higher frequency of western mitochondrial DNA haplotypes (37.5%) than the Xiongnu from the Egyin Gol necropolis (Rogers 2016: 78)."</ref> | |||
| === Paternal lineages === | |||
| === Xianbei people === | |||
| A genetic study published in the ''American Journal of Physical Anthropology'' in August 2018 noted that the paternal ] has been detected among the Xianbei and the ], and was probably an important lineage among the ].{{sfn|Li et al.|2018|p=1}} | |||
| {{Main|Xianbei}} | |||
| === |
==== Autosomal DNA ==== | ||
| A full genome study on multiple Xianbei remains found them to be derived primarily to exclusively from the ] gene pool.<ref>{{Cite journal |last1=Cai |first1=Dawei |last2=Zheng |first2=Ying |last3=Bao |first3=Qingchuan |last4=Hu |first4=Xiaonong |last5=Chen |first5=Wenhu |last6=Zhang |first6=Fan |last7=Cao |first7=Jianen |last8=Ning |first8=Chao |date=24 November 2023 |title=Ancient DNA sheds light on the origin and migration patterns of the Xianbei confederation |url=https://doi.org/10.1007/s12520-023-01899-x |journal=Archaeological and Anthropological Sciences |language=en |volume=15 |issue=12 |pages=194 |doi=10.1007/s12520-023-01899-x |bibcode=2023ArAnS..15..194C |s2cid=265381985 |issn=1866-9565}}</ref> | |||
| Genetic studies published in 2006 and 2015 revealed that the ] of Xianbei remains were of ] origin. According to Zhou (2006) the maternal haplogroup frequencies of the Tuoba Xianbei were 43.75% ], 31.25% ], 12.5% ], 6.25% ] and 6.25% "other." And the inner Mongolia of Tuoba Xianbei<ref>{{Cite journal |vauthors=Changchun Y, Li X, Xiaolei Z, Hui Z, Hong Z |date=November 2006 |title=Genetic analysis on Tuoba Xianbei remains excavated from Qilang Mountain Cemetery in Qahar Right Wing Middle Banner of Inner Mongolia |journal=FEBS Letters |volume=580 |issue=26 |pages=6242–6246 |doi=10.1016/j.febslet.2006.10.030 |pmid=17070809 |ref=Zhou2006 |doi-access=free |s2cid=19492267}}</ref> According to Hong Zhu (2007) The maternal haplogroup frequencies of the Tuoba Xianbei in the Inner Mongolia Autonomous Region were 40.10% Haplogroup O (Y-haplo), 30.16% Haplogroup C (Y-haplo) and 27.94% "other".<ref name="onlinelibrary.wiley.com">{{Cite journal |display-authors=6 |vauthors=Wang H, Ge B, Mair VH, Cai D, Xie C, Zhang Q, Zhou H, Zhu H |date=November 2007 |title=Molecular genetic analysis of remains from Lamadong cemetery, Liaoning, China |journal=American Journal of Physical Anthropology |volume=134 |issue=3 |pages=404–411 |doi=10.1002/ajpa.20685 |pmid=17632796}}</ref> | |||
| Zhou (2014) obtained ] analysis from 17 Tuoba Xianbei, which indicated that these specimens were, similarly, completely East Asian in their maternal origins, belonging to haplogroups D, C, B, A, O and ].<ref name="onlinelibrary.wiley.com"/><ref>{{Cite journal |vauthors=Zhou H |date=March 2014 |title=Genetic analyses of Xianbei populations about 1,500–1,800 years old |journal=Human Genetics |volume=50 |issue=3 |pages=308–314 |doi=10.1134/S1022795414030119 |ref=Zhou2014 |s2cid=18809679}}</ref> | |||
| The research also found a relation between Xianbei individuals with modern ], ] and ]n people. Especially ] Oroqen show close relation to Xianbei.<ref>{{Cite journal |vauthors=Changchun Y, Li X, Xiaolei Z, Hui Z, Hong Z |date=November 2006 |title=Genetic analysis on Tuoba Xianbei remains excavated from Qilang Mountain Cemetery in Qahar Right Wing Middle Banner of Inner Mongolia |journal=FEBS Letters |volume=580 |issue=26 |pages=6242–6 |doi=10.1016/j.febslet.2006.10.030 |pmid=17070809 |doi-access=free |s2cid=19492267}}</ref> | |||
| == Genetic history of Manchus and Daurs == | |||
| === Paternal lineages === | |||
| ]]<ref>{{Cite journal |display-authors=6 |vauthors=Wei LH, Yan S, Yu G, Huang YZ, Yao DL, Li SL, Jin L, Li H |date=March 2017 |title=Genetic trail for the early migrations of Aisin Gioro, the imperial house of the Qing dynasty |journal=Journal of Human Genetics |volume=62 |issue=3 |pages=407–411 |doi=10.1038/jhg.2016.142 |pmid=27853133 |s2cid=7685248}}</ref><ref>{{Cite journal |vauthors=Yan S, Tachibana H, Wei LH, Yu G, Wen SQ, Wang CC |date=June 2015 |title=Y chromosome of Aisin Gioro, the imperial house of the Qing dynasty |journal=Journal of Human Genetics |volume=60 |issue=6 |pages=295–8 |arxiv=1412.6274 |doi=10.1038/jhg.2015.28 |pmid=25833470 |s2cid=7505563}}</ref><ref name="DidYouKnowDNA">{{Cite web |date=14 November 2016 |title=Did you know DNA was used to uncover the origin of the House of Aisin Gioro? |url=https://www.didyouknowdna.com/famous-dna/aisin-gioro-dna/ |access-date=5 November 2020 |website=Did You Know DNA...}}</ref> has been identified as a possible marker of the ] and is found in ten different ethnic minorities in northern China, but completely absent from Han Chinese.<ref>{{Cite journal |display-authors=6 |vauthors=Xue Y, Zerjal T, Bao W, Zhu S, Lim SK, Shu Q, Xu J, Du R, Fu S, Li P, Yang H, Tyler-Smith C |date=December 2005 |title=Recent spread of a Y-chromosomal lineage in northern China and Mongolia |journal=American Journal of Human Genetics |volume=77 |issue=6 |pages=1112–6 |doi=10.1086/498583 |pmc=1285168 |pmid=16380921}}</ref><ref>{{Cite web |title=Asian Ancestry based on Studies of Y-DNA Variation: Part 3. Recent demographics and ancestry of the male East Asians – Empires and Dynasties |url=http://www.genebase.com/learning/article/23 |url-status=dead |archive-url=https://web.archive.org/web/20131125053101/http://www.genebase.com/learning/article/23 |archive-date=25 November 2013 |website=Genebase Tutorials}}</ref><ref name="DidYouKnowDNA"/> | |||
| ] also showed that the haplogroup C3b1a3a2-F8951 of the Aisin Gioro family came to southeastern Manchuria after migrating from their place of origin in the Amur river's middle reaches, originating from ancestors related to ] in the ] area. The ] speaking peoples mostly have C3c-M48 as their subclade of C3 which drastically differs from the C3b1a3a2-F8951 haplogroup of the Aisin Gioro which originates from Mongolic speaking populations like the Daur. Jurchen (Manchus) are a Tungusic people. The Mongol Genghis Khan's haplogroup C3b1a3a1-F3796 (C3*-Star Cluster) is a fraternal "brother" branch of C3b1a3a2-F8951 haplogroup of the Aisin Gioro.<ref>{{Cite journal |display-authors=6 |vauthors=Wei LH, Yan S, Yu G, Huang YZ, Yao DL, Li SL, Jin L, Li H |date=March 2017 |title=Genetic trail for the early migrations of Aisin Gioro, the imperial house of the Qing dynasty |url=https://www.researchgate.net/publication/310477623 |journal=Journal of Human Genetics |publisher=The Japan Society of Human Genetics |volume=62 |issue=3 |pages=407–411 |doi=10.1038/jhg.2016.142 |pmid=27853133 |s2cid=7685248}}</ref> | |||
| A genetic test was conducted on seven men who claimed Aisin Gioro descent with three of them showing documented genealogical information of all their ancestors up to Nurhaci. Three of them turned out to share the C3b2b1*-M401(xF5483) haplogroup, out of them, two of them were the ones who provided their documented family trees. The other four tested were unrelated.<ref>{{Cite journal |vauthors=Yan S, Tachibana H, Wei LH, Yu G, Wen SQ, Wang CC |date=June 2015 |title=Y chromosome of Aisin Gioro, the imperial house of the Qing dynasty |url=https://www.researchgate.net/publication/269876995 |journal=Journal of Human Genetics |publisher=Nature Publishing Group on behalf of the Japan Society of Human Genetics (Japan) |volume=60 |issue=6 |pages=295–8 |arxiv=1412.6274 |doi=10.1038/jhg.2015.28 |pmid=25833470 |s2cid=7505563}}</ref> The Daur Ao clan carries the unique haplogroup subclade C2b1a3a2-F8951, the same haplogroup as Aisin Gioro and both Ao and Aisin Gioro only diverged merely a couple of centuries ago from a shared common ancestor. Other members of the Ao clan carry haplogroups like N1c-M178, C2a1b-F845, C2b1a3a1-F3796 and C2b1a2-M48. People from northeast China, the Daur Ao clan and Aisin Gioro clan are the main carriers of haplogroup C2b1a3a2-F8951. The Mongolic C2*-Star Cluster (C2b1a3a1-F3796) haplogroup is a fraternal branch to Aisin Gioro's C2b1a3a2-F8951 haplogroup.<ref>{{Cite journal |vauthors=Wang CZ, Wei LH, Wang LX, Wen SQ, Yu XE, Shi MS, Li H |date=August 2019 |title=Relating Clans Ao and Aisin Gioro from northeast China by whole Y-chromosome sequencing |journal=Journal of Human Genetics |publisher=Japan Society of Human Genetics |volume=64 |issue=8 |pages=775–780 |doi=10.1038/s10038-019-0622-4 |pmid=31148597 |s2cid=171094135}}</ref> | |||
| However, the modern ] show relatively high amounts of ], which is common among the Han Chinese. A study on the Manchu population of ] reported that they have a close genetic relationship and significant admixture signal with northern ]. The Liaoning Manchu were formed from a major ancestral component related to ] farmers and a minor ancestral component linked to ancient populations from the ] Basin, or others. The Manchu were therefore an exception to the coherent genetic structure of Tungusic-speaking populations, likely due to the large-scale population migrations and genetic admixtures in the past few hundred years.<ref>{{cite journal | vauthors = Zhang X, He G, Li W, Wang Y, Li X, Chen Y, Qu Q, Wang Y, Xi H, Wang CC, Wen Y | display-authors = 6 | title = Genomic Insight Into the Population Admixture History of Tungusic-Speaking Manchu People in Northeast China | journal = Frontiers in Genetics | volume = 12 | pages = 754492 | date = 30 September 2021 | pmid = 34659368 | pmc = 8515022 | doi = 10.3389/fgene.2021.754492 | doi-access = free }} {{CC-notice|cc=by4}}</ref> | |||
| == Genetic history of Japanese == | |||
| {{Main|Genetic and anthropometric studies on Japanese people}} | |||
| === Ainu people === | |||
| Recent research suggests that the historical Ainu culture originated from a merger of the ] with the ], cultures thought to have derived from the diverse Jōmon-period cultures of the Japanese archipelago.<ref name="Sato2007" /><ref>{{cite journal |vauthors=Lee S, Hasegawa T |year=2013 |title=Evolution of the Ainu language in space and time |journal=PLOS ONE |volume=8 |issue=4 |pages=e62243 |bibcode=2013PLoSO...862243L |doi=10.1371/journal.pone.0062243 |pmc=3637396 |pmid=23638014 |doi-access=free}}</ref> According to Lee and Hasegawa of the ], the direct ancestors of the later Ainu people formed during the late ] from the combination of the local but diverse population of ], long before the arrival of contemporary ]. Lee and Hasegawa suggest that the ] expanded from northern Hokkaido and may have originated from a relative more recent Northeast Asian/Okhotsk population, which established themselves in northern Hokkaido and had significant impact on the formation of Hokkaido's Jōmon culture.<ref>{{cite journal |vauthors=Lee S, Hasegawa T |date=April 2013 |title=Evolution of the Ainu language in space and time |journal=PLOS ONE |volume=8 |issue=4 |pages=e62243 |bibcode=2013PLoSO...862243L |doi=10.1371/journal.pone.0062243 |pmc=3637396 |pmid=23638014 |quote=In this paper, we reconstructed spatiotemporal evolution of 19 Ainu language varieties, and the results are in strong agreement with the hypothesis that a recent population expansion of the Okhotsk people played a critical role in shaping the Ainu people and their culture. Together with the recent archaeological, biological and cultural evidence, our phylogeographic reconstruction of the Ainu language strongly suggests that the conventional dual-structure model must be refined to explain these new bodies of evidence. The case of the Ainu language origin we report here also contributes additional detail to the global pattern of language evolution, and our language phylogeny might also provide a basis for making further inferences about the cultural dynamics of the Ainu speakers . |doi-access=free}}</ref><ref>{{Cite journal |vauthors=Matsumoto H |date=February 2009 |title=The origin of the Japanese race based on genetic markers of immunoglobulin G |journal=Proceedings of the Japan Academy. Series B, Physical and Biological Sciences |volume=85 |issue=2 |pages=69–82 |bibcode=2009PJAB...85...69M |doi=10.2183/pjab.85.69 |pmc=3524296 |pmid=19212099}}</ref> | |||
| Recently in 2021, it was confirmed that the Hokkaido Jōmon people formed from "Jōmon tribes of Honshu" and from "Terminal Upper-Paleolithic people" (TUP people) indigenous to Hokkaido and Paleolithic Northern Eurasia. The Honshu Jōmon groups arrived about 15,000 BC and merged with the indigenous "TUP people" to form the Hokkaido Jōmon. The Ainu in turn formed from the Hokkaido Jōmon and from the Okhotsk people.<ref>{{Cite journal |vauthors=Natsuki D |date=19 January 2021 |title=Migration and adaptation of Jomon people during Pleistocene/Holocene transition period in Hokkaido, Japan |journal=Quaternary International |language=en |volume=608–609 |pages=49–64 |doi=10.1016/j.quaint.2021.01.009 |issn=1040-6182 |s2cid=234215606 |quote=The Incipient Jomon communities coexisted with the Terminal Upper Paleolithic (TUP) people that had continued to occupy the region since the stage prior to the LG warm period, but the Incipient Jomon population was relatively small. |doi-access=free}}</ref> | |||
| === Ancient Japan === | |||
| Jōmon people is the generic name of people who lived in the ] during the ]. Today most Japanese historians believe that the Jōmon people were not one homogeneous people but were at least two or three distinct groups.<ref>{{Cite web |date=3 June 2016 |title=The Origins of Japanese Culture Uncovered Using DNA ― What happens when we cut into the world of the Kojiki myths using the latest science |url=https://sp.japanpolicyforum.jp/archives/culture/pt20160603213440.html |url-status=dead |archive-url=https://web.archive.org/web/20190121232846/https://sp.japanpolicyforum.jp/archives/culture/pt20160603213440.html |archive-date=21 January 2019 |access-date=21 January 2019 |website=Discuss Japan-Japan Foreign Policy Forum |vauthors=Miura S, Shinoda K}}</ref> | |||
| Recent full genome analyses in 2020 by Boer et al. 2020 and Yang et al. 2020, reveal some further information regarding the origin of the Jōmon peoples. They were found to have largely formed from a Paleolithic Siberian population and an East Asian related population.<ref name="auto1"/><ref>{{Cite journal |vauthors=de Boer E, Yang MA, Kawagoe A, Barnes GL |date=2020 |title=Japan considered from the hypothesis of farmer/language spread |journal=Evolutionary Human Sciences |language=en |volume=2 |pages=e13 |doi=10.1017/ehs.2020.7 |pmid=37588377 |pmc=10427481 |issn=2513-843X |doi-access=free}}</ref> | |||
| The analyses found that the Jōmon maintained a small effective population size of around 1,000 over several millennia. The Jōmon lineage split from modern East Asians between 15,000 BCE and 20,000 BCE, and became largely isolated from outside populations, but received gene flow from a population related to the Upper-Paleolithic ] sample from Northern Siberia, a deeply European-related population, also known as ]. Niall Cooke, one of the researchers explained that these results strongly suggest a prolonged period of isolation from the rest of the continent until the introduction of new immigrants associated with wet-rice farming during the Yayoi period of Japanese history.<ref name="Niall22"/> | |||
| The ] were migrants to the ] from ] (] or ]) during the ] (1000 BCE–300 CE) and ] (250–538 CE). They are seen as direct ancestors of the modern ], the majority of Japanese and of the ]. It is estimated that modern Japanese share in average about 90% of their genome with the Yayoi.<ref>{{cite web |title='Jomon woman' helps solve Japan's genetic mystery {{!}} NHK WORLD-JAPAN News |url=https://www3.nhk.or.jp/nhkworld/en/news/backstories/555/ |access-date=9 July 2019 |website=NHK WORLD |language=en}}</ref> | |||
| This group is thought to have marked the arrival of ] rice cultivation which led to an agricultural revolution in the archipelago until the following period.<ref name="Niall22"/> | |||
| A study published in the journal ]<ref name=":12">{{Cite web |last=Trinity College Dublin |date=17 September 2021 |title=Ancient DNA rewrites early Japanese history—modern day populations have tripartite genetic origin |url=https://phys.org/news/2021-09-ancient-dna-rewrites-early-japanese.html |access-date=1 March 2022 |website=phys.org |language=en}}</ref> found that the people of Japan bore genetic signatures from three ancient populations rather than just two as previously thought, with ancestry stemming from a third migration that occurred during the ].<ref name="Niall22">{{cite journal | vauthors = Cooke NP, Mattiangeli V, Cassidy LM, Okazaki K, Stokes CA, Onbe S, Hatakeyama S, Machida K, Kasai K, Tomioka N, Matsumoto A, Ito M, Kojima Y, Bradley DG, Gakuhari T, Nakagome S | display-authors = 6 | title = Ancient genomics reveals tripartite origins of Japanese populations | journal = Science Advances | volume = 7 | issue = 38 | pages = eabh2419 | date = September 2021 | pmid = 34533991 | pmc = 8448447 | doi = 10.1126/sciadv.abh2419 | bibcode = 2021SciA....7.2419C }}</ref> The study found that up to 71% of the ancient Kofun people shared a common genetic strand with the ] while the rest shared with the ] and the ]. In addition, '']'' published an article that showed the Kofun strand in modern day Japanese was concentrated in specific regions such as ], ] and ].<ref name=":022">{{Cite web |last=Nikkei Science |date=23 June 2021 |title=渡来人、四国に多かった? ゲノムが明かす日本人ルーツ |trans-title=Were there many migrants in Shikoku? Japanese roots revealed by genome analysis |url=https://www.nikkei.com/article/DGXZQOUC18CCA0Y1A610C2000000/ |access-date=1 May 2022 |website=nikkei.com |language=ja}}</ref> | |||
| There is evidence of population admixture in ancient Korea. A 2022 study conducted by the ] found that an ancient Korea had considerable genetic diversity, including elevated levels of Jomon ancestry.<ref name=":4">{{cite journal |last1=Wang |first1=Rui |last2=Wang |first2=Chuan-Chao |date=8 August 2022 |title=Human genetics: The dual origin of Three Kingdoms period Koreans |journal=Current Biology |language=en |volume=32 |issue=15 |pages=R844–R847 |doi=10.1016/j.cub.2022.06.044 |pmid=35944486 |s2cid=251410856 |issn=0960-9822 |doi-access=free}} "Present-day Koreans are closely genetically related to the Three Kingdoms period individuals inferred from allele and haplotype sharing patterns and the profiles of shared dominant Y-chromosome and mitochondrial DNA haplogroups. Several specific phenotypic trait-related genetic variants of present-day Koreans were already present in the ancient genomes of the Three Kingdoms period8. Although ancient Koreans with Jomon ancestry contributed substantially to the genetic formation of present-day Koreans, the present-day Koreans show high genetic homogeneity without Jomon-related ancestry. Whether the Bronze Age Korean individual from Taejungni had been genetically similar to present-day Korean individuals was not convincingly shown due to the very low genomic coverage5. Taking no account of this Bronze Age sample, the possible scenario was that the indigenous Jomon-related ancestry was largely replaced through admixture with the incoming northern China populations and disappeared in the later centuries, creating relatively homogeneous present-day Koreans."</ref> It was hypothesized that the Jomon ancestry of ancient Koreans was lost over time, as they continually mixed with incoming populations from northern China, followed by a period of isolation during the Three Kingdoms period, resulting in the homogenous gene pool of modern Koreans.<ref name=":4" /><ref name=":1">{{cite web |title=1,700-year-old Korean genomes show genetic heterogeneity in Three Kingdoms period Gaya |url=https://www.sciencedaily.com/releases/2022/06/220621114705.htm |access-date=29 August 2022 |website=ScienceDaily |language=en}}</ref> | |||
| === Modern Japanese === | |||
| ]'s cluster (square) is almost indistinguishable to the ]'s cluster (circle), while the Jōmon samples are shifted towards the Siberian cluster in a more distinct position. (2019)]]According to a study on genetic distance measurements from a large scale genetic study from 2021 titled 'Genomic insights into the formation of human populations in East Asia', the modern "Japanese populations can be modelled as deriving from Korean (91%) and Jōmon (9%)."<ref>{{Cite journal |last=Wang |first=Chuan-Chao |date=2021 |title=Genomic insights into the formation of human populations in East Asia |url=https://www.researchgate.net/publication/349510968 |journal=Nature |volume=591 |issue=7850 |pages=413–419 |bibcode=2021Natur.591..413W |doi=10.1038/s41586-021-03336-2 |pmc=7993749 |pmid=33618348}}</ref>] (classified under the Japanese-Korean cluster) with other East Asians. (2022)]] | |||
| ] | |||
| ==== Paternal lineages ==== | ==== Paternal lineages ==== | ||
| A genetic study published in the ''American Journal of Physical Anthropology'' in August 2018 noted that the paternal ] has been detected among the Xianbei and the ], and was probably an important lineage among the ].{{sfn|Li et al.|2018|p=1}} | |||
| A comprehensive study of worldwide Y-DNA diversity (Underhill ''et al.'' 2000) included a sample of 23 males from Japan, of whom 35% belonged to ], 26% belonged to ], 22% belonged to ], 13% belonged to ] and ], and 4.3% belonged to ].<ref name="Underhill_2000">{{cite journal |display-authors=6 |vauthors=Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonné-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi SQ, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza LL, Oefner PJ |date=November 2000 |title=Y chromosome sequence variation and the history of human populations |journal=Nature Genetics |volume=26 |issue=3 |pages=358–61 |doi=10.1038/81685 |pmid=11062480 |s2cid=12893406}}</ref> | |||
| Poznik ''et al.'' (2016) reported the haplogroups of a sample of Japanese men from Tokyo:<ref>The JPT sample is considered "as generally representative of the majority population in Japan". See {{Cite web |title=Japanese in Tokyo, Japan – Population Description |url=https://catalog.coriell.org/0/Sections/Collections/NHGRI/Japanese.aspx?PgId=359 |publisher=Coriell Institute for Medical Research |location=Camden, ] |vauthors=Matsuda I}}</ref> 36% belonged to ], 32% had ], 18% carried ], 7.1% carried ], 3.6% belonged to ], and 3.6% carried ].<ref name="Poznik2016">{{Cite journal |display-authors=6 |vauthors=Poznik GD, Xue Y, Mendez FL, Willems TF, Massaia A, Wilson Sayres MA, Ayub Q, McCarthy SA, Narechania A, Kashin S, Chen Y, Banerjee R, Rodriguez-Flores JL, Cerezo M, Shao H, Gymrek M, Malhotra A, Louzada S, Desalle R, Ritchie GR, Cerveira E, Fitzgerald TW, Garrison E, Marcketta A, Mittelman D, Romanovitch M, Zhang C, Zheng-Bradley X, Abecasis GR, McCarroll SA, Flicek P, Underhill PA, Coin L, Zerbino DR, Yang F, Lee C, Clarke L, Auton A, Erlich Y, Handsaker RE, Bustamante CD, Tyler-Smith C |date=June 2016 |title=Punctuated bursts in human male demography inferred from 1,244 worldwide Y-chromosome sequences |journal=Nature Genetics |volume=48 |issue=6 |pages=593–9 |doi=10.1038/ng.3559 |pmc=4884158 |pmid=27111036 |hdl=11858/00-001M-0000-002A-F024-C}}</ref> | |||
| ==== Maternal lineages ==== | ==== Maternal lineages ==== | ||
| Genetic studies published in 2006 and 2015 revealed that the ] of Xianbei remains were of ] origin. According to Zhou (2006) the maternal haplogroup frequencies of the Tuoba Xianbei were 43.75% ], 31.25% ], 12.5% ], 6.25% ] and 6.25% "other".<ref>{{Cite journal |vauthors=Changchun Y, Li X, Xiaolei Z, Hui Z, Hong Z |date=November 2006 |title=Genetic analysis on Tuoba Xianbei remains excavated from Qilang Mountain Cemetery in Qahar Right Wing Middle Banner of Inner Mongolia |journal=FEBS Letters |volume=580 |issue=26 |pages=6242–6246 |doi=10.1016/j.febslet.2006.10.030 |pmid=17070809 |ref=Zhou2006 |doi-access=free |bibcode=2006FEBSL.580.6242C |s2cid=19492267}}</ref> Zhou (2014) obtained ] analysis from 17 Tuoba Xianbei, which indicated that these specimens were, similarly, completely East Asian in their maternal origins, belonging to haplogroups D, C, B, A, O and ].<ref>{{Cite journal |display-authors=6 |vauthors=Wang H, Ge B, Mair VH, Cai D, Xie C, Zhang Q, Zhou H, Zhu H |date=November 2007 |title=Molecular genetic analysis of remains from Lamadong cemetery, Liaoning, China |journal=American Journal of Physical Anthropology |volume=134 |issue=3 |pages=404–411 |doi=10.1002/ajpa.20685 |pmid=17632796 |doi-access=free}}</ref><ref>{{Cite journal |vauthors=Zhou H |date=March 2014 |title=Genetic analyses of Xianbei populations about 1,500–1,800 years old |journal=Human Genetics |volume=50 |issue=3 |pages=308–314 |doi=10.1134/S1022795414030119 |ref=Zhou2014 |s2cid=18809679}}</ref> | |||
| According to an analysis of the ]'s sample of Japanese collected in the Tokyo metropolitan area, the mtDNA haplogroups found among modern Japanese include ] (42/118 = 35.6%, including 39/118 = 33.1% D4 and 3/118 = 2.5% D5), ] (16/118 = 13.6%, including 11/118 = 9.3% B4 and 5/118 = 4.2% B5), ] (12/118 = 10.2%), ] (12/118 = 10.2%), ] (10/118 = 8.5%), ] (9/118 = 7.6%), ] (8/118 = 6.8%), ] (4/118 = 3.4%), ] (3/118 = 2.5%), and ] (2/118 = 1.7%).<ref name="Zheng_2011">{{Cite journal |vauthors=Zheng HX, Yan S, Qin ZD, Wang Y, Tan JZ, Li H, Jin L |year=2011 |title=Major population expansion of East Asians began before neolithic time: evidence of mtDNA genomes |journal=PLOS ONE |volume=6 |issue=10 |pages=e25835 |bibcode=2011PLoSO...625835Z |doi=10.1371/journal.pone.0025835 |pmc=3188578 |pmid=21998705 |doi-access=free}}</ref> | |||
| === Jōmon people === | |||
| {{Main|Jōmon people}} | |||
| A 2011 SNP consortium study done by the ] and ] consisting of 1719 DNA samples determined that Koreans and Japanese clustered near to each other, confirming the findings of an earlier study that Koreans and Japanese are related.<ref>{{cite journal | vauthors = Yang X, Xu S, ((HUGO Pan-Asian SNP Consortium)), ((Indian Genome Variation Consortium)) | title = Identification of close relatives in the HUGO Pan-Asian SNP database | journal = PLOS ONE | volume = 6 | issue = 12 | pages = e29502 | date = 29 December 2011 | pmid = 22242128 | pmc = 3248454 | doi = 10.1371/journal.pone.0029502 | bibcode = 2011PLoSO...629502Y | doi-access = free }}</ref> | |||
| The Jōmon people represent the indigenous population of the ] during the ]. They are inferred to descend from the Paleolithic inhabitants of ]. Genetic analyses on Jōmon remains found them to represent a deeply diverged ] lineage. The Jōmon lineage is inferred to have diverged from Ancient East Asians before the divergence between ]s and ]s, but after the divergence of the basal ] and or ]s. Beyond their broad affinity with Eastern Asian lineages, the Jōmon also display a weak affinity for ]s (ANE), which may be associated with the introduction of ] to Northeast Asia and northern East Asia during the ] via the ANE or ]s.<ref>{{Cite journal |last1=Osada |first1=Naoki |last2=Kawai |first2=Yosuke |date=2021 |title=Exploring models of human migration to the Japanese archipelago using genome-wide genetic data |url=https://www.jstage.jst.go.jp/article/ase/129/1/129_201215/_html/-char/en |journal=Anthropological Science |volume=129 |issue=1 |pages=45–58 |doi=10.1537/ase.201215 |doi-access=free}}</ref><ref name=":92" /> | |||
| A 2008 study about genome-wide SNPs of East Asians by Chao Tian ''et al.'' reported that Japanese along with other East Asians such as ] and ] are genetically distinguishable from Southeast Asians<ref name="Public Library of Science">{{cite journal | vauthors = Tian C, Kosoy R, Lee A, Ransom M, Belmont JW, Gregersen PK, Seldin MF | title = Analysis of East Asia genetic substructure using genome-wide SNP arrays | journal = PLOS ONE | volume = 3 | issue = 12 | pages = e3862 | date = 5 December 2008 | pmid = 19057645 | pmc = 2587696 | doi = 10.1371/journal.pone.0003862 | bibcode = 2008PLoSO...3.3862T | doi-access = free }}</ref> Another study (2017) shows a relative strong relation between all East and Southeast Asians.<ref>{{Cite web|url=http://mengnews.joins.com/view.aspx?aId=3030017|title=Pinning down Korean-ness through DNA|website=Korea JoongAng Daily|language=ko|access-date=9 July 2019}}</ref> | |||
| === Hoabinhians === | |||
| == Genetic history of Koreans == | |||
| {{Main|Hoabinhian}}The Hoabinhians represent a technologically advanced society of hunter-gatherers, primarily living in ], but also adjacent regions of ]. While the Upper Paleolithic origins of this 'Hoabinhian ancestry' are unknown, Hoabinhian ancestry has been found to be related to the main 'East Asian' ancestry component found in most modern East and Southeast Asians, although deeply diverged from it.<ref>{{cite journal |last1=McColl |first1=Hugh |last2=Racimo |first2=Fernando |last3=Vinner |first3=Lasse |last4=Demeter |first4=Fabrice |last5=Gakuhari |first5=Takashi |last6=Moreno-Mayar |first6=J. Víctor |last7=van Driem |first7=George |last8=Gram Wilken |first8=Uffe |last9=Seguin-Orlando |first9=Andaine |last10=de la Fuente Castro |first10=Constanza |last11=Wasef |first11=Sally |last12=Shoocongdej |first12=Rasmi |last13=Souksavatdy |first13=Viengkeo |last14=Sayavongkhamdy |first14=Thongsa |last15=Saidin |first15=Mohd Mokhtar |display-authors=5 |year=2018 |title=The prehistoric peopling of Southeast Asia |journal=Science |publisher=American Association for the Advancement of Science (AAAS) |volume=361 |issue=6397 |pages=88–92 |bibcode=2018Sci...361...88M |doi=10.1126/science.aat3628 |issn=0036-8075 |pmid=29976827 |s2cid=206667111 |hdl-access=free |last16=Allentoft |first16=Morten E. |last17=Sato |first17=Takehiro |last18=Malaspinas |first18=Anna-Sapfo |last19=Aghakhanian |first19=Farhang A. |last20=Korneliussen |first20=Thorfinn |hdl=10072/383365|url=https://research.monash.edu/en/publications/5420ab64-ae26-43a7-98dc-9d08834807fc }}</ref><ref>{{Cite journal |last=Yang |first=Melinda A. |date=6 January 2022 |title=A genetic history of migration, diversification, and admixture in Asia |journal=Human Population Genetics and Genomics |volume=2 |issue=1 |pages=1–32 |doi=10.47248/hpgg2202010001 |doi-access=free}}</ref> Together with the Paleolithic ], they form early branches of East Asian genetic diversity, and are described as "Basal Asian" (BA) or "Basal East Asian" (BEA).<ref>{{Cite journal |last1=张明 |first1=平婉菁 |last2=ZHANG Ming |first2=PING Wanjing |date=15 June 2023 |title=古基因组揭示史前欧亚大陆现代人复杂遗传历史 |url=https://www.anthropol.ac.cn/CN/10.16359/j.1000-3193/AAS.2023.0010 |journal=人类学学报 |language=zh |volume=42 |issue=3 |pages=412 |doi=10.16359/j.1000-3193/AAS.2023.0010 |issn=1000-3193}}</ref> | |||
| == Modern populations == | |||
| ]{{Main|East Asian peoples}} | |||
| Studies of ] have so far produced evidence to suggest that the ] have a long history as a distinct, mostly ] ethnic group, with successive waves of people moving to the peninsula and three major Y-chromosome haplogroups.<ref>{{Cite journal |vauthors=Kim SH, Han MS, Kim W, Kim W |date=November 2010 |title=Y chromosome homogeneity in the Korean population |journal=International Journal of Legal Medicine |volume=124 |issue=6 |pages=653–7 |doi=10.1007/s00414-010-0501-1 |pmid=20714743 |s2cid=27125545}}</ref> The reference population for Koreans used in ] is 94% Eastern Asia and 5% Southeast Asia & Oceania.<ref>]. (2017). ]. Retrieved 15 May 2017, from </ref> | |||
| ] Korean genomes. (Gelabert 2022)]] | |||
| Veronika Siska et al. (2017) said that the ] are genetically closest in the study's panel to the human remains from the ] which are dated to about 7,700 years ago.<!--The 7,700 date comes from the second sentence of the "Results" section on page 1 of 10. In that sentence, it's written as "~7.7 ka". In the third sentence of the article's abstract on page 1 of 10, the date is alternatively written as "~7.7 thousand years ago".--> Modern Korean and Japanese, the ] and the ] display a high affinity to the human remains from Devil's Gate Cave. Considering the geographic distance of ] from Devil's Gate Cave, Amerindians are unusually genetically close to the human remains from Devil's Gate Cave. Korean ]s display similar traits to Japanese genomes on genome-wide ] data. In an admixture analysis, when the genes of Devil's Gate are made into a unique genetic component, this new Devil's Gate genetic component is highest in peoples of the Amur Basin, including Ulchi, and makes up more than 50% of Koreans and Japanese. It also has a sporadic distribution among other East Asians, Central Asians and Southeast Asians.<ref name="pmid28164156">{{Cite journal |display-authors=6 |vauthors=Siska V, Jones ER, Jeon S, Bhak Y, Kim HM, Cho YS, Kim H, Lee K, Veselovskaya E, Balueva T, Gallego-Llorente M, Hofreiter M, Bradley DG, Eriksson A, Pinhasi R, Bhak J, Manica A |date=February 2017 |title=Genome-wide data from two early Neolithic East Asian individuals dating to 7700 years ago |journal=Science Advances |volume=3 |issue=2 |pages=e1601877 |bibcode=2017SciA....3E1877S |doi=10.1126/sciadv.1601877 |pmc=5287702 |pmid=28164156}}</ref> | |||
| ] | |||
| === Manchu and Daur peoples === | |||
| According to a study conducted in 2022, ancient samples from southern Korea had considerably higher ] ancestry than modern Korean people. All samples could be modeled as a mixture of two components: northern China Bronze Age-related ancestry and Jomon-related ancestry. The authors said that this elevated Jomon ancestry persisted in the Korean peninsula until at least the year ].<ref name=":7">{{Cite journal |last=Gelabert |first=Pere |date=2022-08-08 |title=Northeastern Asian and Jomon-related genetic structure in the Three Kingdoms period of Gimhae, Korea |url=https://www.sciencedirect.com/science/article/pii/S0960982222009162 |journal=Current Biology |volume=32 |issue=15 |pages=3232–3244.e6 |doi=10.1016/j.cub.2022.06.004 |issn=0960-9822}}</ref><ref>{{harvnb|Gelabert|2022|ps=: Similarly, we demonstrated that the reported lack of Jomon-related ancestry in the previously published single Bronze Age Korean genome may not be statistically supported with such low amounts of data10 and with the analytical tools used (i.e., qpAdm). Nevertheless, our results show that Jomon-related ancestry persisted at high levels in South Korea until at least the TK period, around 500 CE."}}</ref> All of the ancient specimens carried ] haplogroups that are common in modern Koreans, but absent in ancient Jomon. Of three males sequenced, two belonged to the ] ] and O*, common in modern Korean men, while one belonged to paternal haplogroup ], which is common in Japan.<ref>{{harvnb|Gelabert|2022|ps=: "All individuals had typical East Asian mitochondrial DNA (mtDNA) haplogroups (Table 1; Data S1A), D (n = 5), B, F, and M, when we determined the haplogroups from the consensus. All of these prevail among present-day Koreans,9,12 with the most common being haplogroup D4, which we identified in four of the eight AKGs. This haplogroup was also common among ancient Japanese Yayoi farmers but was absent in the Jomon.32 Out of the three male individuals, we successfully called Y chromosome haplogroups (Table 1; Data S1A) for two: AKG_10203 (D1a2a1) and AKG_10204 (O1b2a1a2a1b1). The third male, AKG_10218, due to his lower coverage, was assigned to the major haplogroup O (Table 1; Data S1A). Haplogroup O is the most common Y haplogroup in present-day Korea and is shared by more than 73% of Korean males,9 whereas haplogroup D is more common in the present-day Japanese population.33"}}</ref> The authors found no relationship between genetic ancestry and the social status of the sampled specimens, and they also found genetic continuity between the ancient Jomon-admixed specimens and modern Koreans, for genes related to hair and eye color, ], and facial morphology.<ref>{{harvnb|Gelabert|2022|ps=: "We could not correlate the genomic differences between these two groups with either social status or sex. All the ancient individuals’ genomic profiles, including phenotypically relevant SNPs associated with hair and eye color, facial morphology, and myopia, imply strong genetic and phenotypic continuity with modern Koreans for the last 1,700 years."}}</ref> | |||
| {{Main|Tungusic peoples}} | |||
| ==== Autosomal DNA ==== | |||
| Of ancient samples dating from the Neolithic era to the Three Kingdoms period in Korea, all but one has some degree of Jōmon ancestry.<ref name=":8">{{Cite web |title=Re-thinking Jōmon and Ainu in Japanese History |url=https://apjjf.org/2022/15/Hudson.html |access-date= |website=The Asia-Pacific Journal: Japan Focus}}</ref> | |||
| A study on the Manchu population of ] reported that they have a close genetic relationship and significant admixture signals from northern ]. The Liaoning Manchu were formed from a major ancestral component related to ] farmers and a minor ancestral component linked to ancient populations from the ] Basin, or others. The Manchu were therefore an exception to the coherent genetic structure of Tungusic-speaking populations, likely due to the large-scale population migrations and genetic admixtures in the past few hundred years.<ref>{{cite journal |display-authors=6 |vauthors=Zhang X, He G, Li W, Wang Y, Li X, Chen Y, Qu Q, Wang Y, Xi H, Wang CC, Wen Y |date=30 September 2021 |title=Genomic Insight Into the Population Admixture History of Tungusic-Speaking Manchu People in Northeast China |journal=Frontiers in Genetics |volume=12 |pages=754492 |doi=10.3389/fgene.2021.754492 |pmc=8515022 |pmid=34659368 |doi-access=free}} {{CC-notice|cc=by4}}</ref> | |||
| ==== Paternal lineages ==== | |||
| A 2022 study was unabld to detect significant Jomon ancestry in modern Koreans, however by using different proxies of ancestry, a Jomon contribution of 3.1-4.4% was found for present-day Ulsan Koreans. Nevertheless, the authors suggested that the model that yielded this result is not the most reliable.<ref>{{cite journal |last1=Lee |first1=Don‐Nyeong |last2=Jeon |first2=Chae Lin |last3=Kang |first3=Jiwon |last4=Burri |first4=Marta |last5=Krause |first5=Johannes |last6=Woo |first6=Eun Jin |last7=Jeong |first7=Choongwon |title=Genomic detection of a secondary family burial in a single jar coffin in early Medieval Korea |journal=American Journal of Biological Anthropology |date=December 2022 |volume=179 |issue=4 |pages=585–597 |doi=10.1002/ajpa.24650 |url=https://onlinelibrary.wiley.com/doi/10.1002/ajpa.24650 |language=en |issn=2692-7691}} "In both ancient and present-day Koreans, we do not detect a statistically significant contribution from the Jomon hunter-gatherer gene pool of the Japanese archipelago (Table S7A), although previous studies report occasional presence of the Jomon ancestry contribution from Neolithic to the early Medieval period (Gelabert et al., 2022; Robbeets et al., 2021). When we replace the genetic northern proxy from WLR_BA to Middle Neolithic individuals from the Miaogizou site in Inner Mongolia (“Miaozigou_MN”), we detect a small but significant amount of Jomon contribution in the Gunsan individuals and present-day Ulsan Koreans (3.1%–4.4%; Table S7B). We believe that WLR_BA provides a more suitable model for ancient and present-day Koreans given its geographical and temporal proximity to them. The remaining well-fitting source pairs provide qualitatively similar results (Table S8)."</ref> | |||
| A plurality of ] males belong to ] (12/39 = 30.8% according to Xue Yali ''et al.'' 2006,<ref name="Xue2006" /> 88/207 = 42.5% according to Wang Chi-zao ''et al.'' 2018<ref name="Wang2018" />), with ] being the second most common haplogroup among present-day Daurs (10/39 = 25.6%,<ref name="Xue2006" /> 52/207 = 25.1%<ref name="Wang2018" />). There are also tribes (''hala''; ''cf.'' ]) among the Daurs that belong predominantly to other Y-DNA haplogroups, such as ] (''Merden hala'') and ] (''Gobulo hala'').<ref name="Wang2018" /> ]<ref>{{Cite journal |display-authors=6 |vauthors=Wei LH, Yan S, Yu G, Huang YZ, Yao DL, Li SL, Jin L, Li H |date=March 2017 |title=Genetic trail for the early migrations of Aisin Gioro, the imperial house of the Qing dynasty |journal=Journal of Human Genetics |volume=62 |issue=3 |pages=407–411 |doi=10.1038/jhg.2016.142 |pmid=27853133 |s2cid=7685248}}</ref><ref>{{Cite journal |vauthors=Yan S, Tachibana H, Wei LH, Yu G, Wen SQ, Wang CC |date=June 2015 |title=Y chromosome of Aisin Gioro, the imperial house of the Qing dynasty |journal=Journal of Human Genetics |volume=60 |issue=6 |pages=295–8 |arxiv=1412.6274 |doi=10.1038/jhg.2015.28 |pmid=25833470 |s2cid=7505563}}</ref><ref name="DidYouKnowDNA" /> has been identified as a possible marker of the ] and is found in ten different ethnic minorities in northern China, but is less prevalent from Han Chinese.<ref>{{Cite journal |display-authors=6 |vauthors=Xue Y, Zerjal T, Bao W, Zhu S, Lim SK, Shu Q, Xu J, Du R, Fu S, Li P, Yang H, Tyler-Smith C |date=December 2005 |title=Recent spread of a Y-chromosomal lineage in northern China and Mongolia |journal=American Journal of Human Genetics |volume=77 |issue=6 |pages=1112–6 |doi=10.1086/498583 |pmc=1285168 |pmid=16380921}}</ref><ref>{{Cite web |title=Asian Ancestry based on Studies of Y-DNA Variation: Part 3. Recent demographics and ancestry of the male East Asians – Empires and Dynasties |url=http://www.genebase.com/learning/article/23 |url-status=dead |archive-url=https://web.archive.org/web/20131125053101/http://www.genebase.com/learning/article/23 |archive-date=25 November 2013 |website=Genebase Tutorials}}</ref><ref name="DidYouKnowDNA" /> The ] also display a significant amount of ], but the most often observed Y-DNA haplogroup among present-day Manchus is ], which they share in common with ].<ref>{{Cite journal |last1=Zhang |first1=Xianpeng |last2=He |first2=Guanglin |last3=Li |first3=Wenhui |last4=Wang |first4=Yunfeng |last5=Li |first5=Xin |last6=Chen |first6=Ying |last7=Qu |first7=Quanying |last8=Wang |first8=Ying |last9=Xi |first9=Huanjiu |last10=Wang |first10=Chuan-Chao |last11=Wen |first11=Youfeng |date=30 September 2021 |title=Genomic Insight Into the Population Admixture History of Tungusic-Speaking Manchu People in Northeast China |journal=Frontiers in Genetics |volume=12 |pages=754492 |doi=10.3389/fgene.2021.754492 |issn=1664-8021 |pmc=8515022 |pmid=34659368 |doi-access=free}}</ref><ref name="Xue2006" /><ref>{{cite journal |last1=Hammer |first1=Michael F. |last2=Karafet |first2=Tatiana M. |last3=Park |first3=Hwayong |last4=Omoto |first4=Keiichi |last5=Harihara |first5=Shinji |last6=Stoneking |first6=Mark |last7=Horai |first7=Satoshi |title=Dual origins of the Japanese: common ground for hunter-gatherer and farmer Y chromosomes |journal=Journal of Human Genetics |date=January 2006 |volume=51 |issue=1 |pages=47–58 |doi=10.1007/s10038-005-0322-0 |pmid=16328082 |s2cid=6559289 |doi-access=free}}</ref> | |||
| === |
=== Ainu people === | ||
| {{Main|Ainu people}} | |||
| ] | |||
| The exact origins of the early Ainu remains unclear, but it is generally agreed to be linked to the ] of the ], with later influences from the nearby ].<ref>{{Cite web |title=公益財団法人 アイヌ民族文化財団 |url=https://www.ff-ainu.or.jp/ |access-date=8 December 2023 |website=www.ff-ainu.or.jp |language=ja}}</ref> The Ainu appear genetically most closely related to the ] of Japan. The genetic makeup of the Ainu represents a "deep branch of East Asian diversity". Compared to contemporary East Asian populations, the Ainu share "a closer genetic relationship with northeast Siberians".<ref name=":2" /><ref name=":92" /> | |||
| Modern Koreans are overall more similar to northeast Asians than to southeast Asians. This conclusion would be expected from the general correlation between genetic variation and geography observed for human populations, and is supported by an examination of individual mtDNA haplogroups, genetic distances between populations derived from mtDNA or Y-chromosomal data, and the apportionment of genetic diversity between different groups of populations.<ref name="Jin" /> | |||
| === Japanese people === | |||
| Studies of classical genetic markers showed that Koreans tend to have a close genetic affinity with Mongolians among East Asians. Ancient genome comparisons revealed that the genetic makeup of Koreans can be best described as an admixture of an Northern hunter-gatherer component as well as that of a influx of rice-farming agriculturalists from the Yangtze river valley, which in turn are often linked to O2-M122 and is largely male-biased.<ref name="Jin" /> Another study concludes however that O2b*-SRY465 and O2b1-47z had an ''in situ'' origin among Northeast Asians, particularly among the prehistoric Koreans, rather than in southern China or Southeast Asia as previously envisaged. The combination of the O2b initial settlement (which became an indigenous proto-Korean component) in part with the relatively recent O3 and C3 lineages (which include a Chinese component) explains some of the main events formulating the current Y chromosome composition of the Korean population.<ref name="Kim2011" /> This is supported by archeological, historical and linguistic evidence, which suggest that the direct ancestors of Koreans were proto-Koreans who inhabited the northeastern region of China and the Korean Peninsula during the Neolithic (8,000–1,000 BC) and Bronze (1,500–400 BC) Ages.<ref name="Kim2011" /> The results from the findings in the Devil's Gate showed that the ancient populations of the area were already admixed from various sources.<ref name=":3">{{cite journal | vauthors = Siska V, Jones ER, Jeon S, Bhak Y, Kim HM, Cho YS, Kim H, Lee K, Veselovskaya E, Balueva T, Gallego-Llorente M, Hofreiter M, Bradley DG, Eriksson A, Pinhasi R, Bhak J, Manica A | display-authors = 6 | title = Genome-wide data from two early Neolithic East Asian individuals dating to 7700 years ago | journal = Science Advances | volume = 3 | issue = 2 | pages = e1601877 | date = February 2017 | pmid = 28164156 | pmc = 5287702 | doi = 10.1126/sciadv.1601877 | bibcode = 2017SciA....3E1877S }}</ref> | |||
| {{Main|Japanese people|Genetic and anthropometric studies on Japanese people}} | |||
| ] of Ainu, Ryukyuan, Mainland Japanese, and other Asian ethnic groups. The Ainu and the Ryukyuan were clustered with 100% bootstrap probability, followed by the Mainland Japanese. The three populations in the Japanese Archipelago clustered with the Korean with 100% bootstrap probability.<ref name="Timothy2012">{{cite journal |last1=Jinam |first1=Timothy |last2=Nishida |first2=Nao |last3=Hirai |first3=Momoki |last4=Kawamura |first4=Shoji |last5=Oota |first5=Hiroki |last6=Umetsu |first6=Kazuo |last7=Kimura |first7=Ryosuke |last8=Ohashi |first8=Jun |last9=Tajima |first9=Atsushi |date=December 2012 |title=The history of human populations in the Japanese Archipelago inferred from genome-wide SNP data with a special reference to the Ainu and the Ryukyuan populations |journal=] |volume=57 |issue=12 |pages=787–795 |doi=10.1038/jhg.2012.114 |pmid=23135232 |doi-access=free}}</ref><ref>{{Cite web |title=記者会見「日本列島3人類集団の遺伝的近縁性」 |url=https://www.u-tokyo.ac.jp/focus/ja/press/p01_241101.html |accessdate=8 November 2021 |website=東京大学 |language=ja}}</ref>|265x265px]] | |||
| Japanese populations in modern Japan can be traced to three separate, but related demographics: the ], ] and Mainland Japanese (]). The populations are closely related to clusters found in Northeastern Asia<ref>{{cite book |author=Mitsuru Sakitani |title=『DNA・考古・言語の学際研究が示す新・日本列島史』 |date=2009 |publisher=Bensei Publishing |isbn=9784585053941 |language=ja |trans-title=New History of the Japanese Islands Shown by Interdisciplinary Studies on DNA, Archeology, and Language}}</ref><ref name="Suzuki2" /><ref>{{cite journal |display-authors=6 |vauthors=Cooke NP, Mattiangeli V, Cassidy LM, Okazaki K, Stokes CA, Onbe S, Hatakeyama S, Machida K, Kasai K, Tomioka N, Matsumoto A, Ito M, Kojima Y, Bradley DG, Gakuhari T, Nakagome S |date=September 2021 |title=Ancient genomics reveals tripartite origins of Japanese populations |journal=Science Advances |volume=7 |issue=38 |pages=eabh2419 |bibcode=2021SciA....7.2419C |doi=10.1126/sciadv.abh2419 |pmc=8448447 |pmid=34533991 |ref={{harvid|Cooke|2021}}}}</ref> with the Ainu group being most similar to the Ryukyuan group,<ref name="Suzuki2" /><ref>{{Cite web |title='Jomon woman' helps solve Japan's genetic mystery {{!}} NHK WORLD-JAPAN News |url=https://www3.nhk.or.jp/nhkworld/en/news/backstories/555/ |access-date=19 November 2020 |website=NHK WORLD |language=en}}</ref> the Ryukyuan group being most similar to the Yamato group,<ref>{{Cite journal |last=Jinam |first=Timothy |last2=Nishida |first2=Nao |last3=Hirai |first3=Momoki |last4=Kawamura |first4=Shoji |last5=Oota |first5=Hiroki |last6=Umetsu |first6=Kazuo |last7=Kimura |first7=Ryosuke |last8=Ohashi |first8=Jun |last9=Tajima |first9=Atsushi |last10=Yamamoto |first10=Toshimichi |last11=Tanabe |first11=Hideyuki |last12=Mano |first12=Shuhei |last13=Suto |first13=Yumiko |last14=Kaname |first14=Tadashi |last15=Naritomi |first15=Kenji |date=8 November 2012 |title=The history of human populations in the Japanese Archipelago inferred from genome-wide SNP data with a special reference to the Ainu and the Ryukyuan populations |url=https://www.nature.com/articles/jhg2012114 |journal=Journal of Human Genetics |language=en |volume=57 |issue=12 |pages=787–795 |doi=10.1038/jhg.2012.114 |issn=1435-232X}}</ref> and the Yamato group being most similar to ]<ref>{{cite AV media |url=https://www.nhk-ondemand.jp/goods/G2018087565SA000/ |title=弥生人DNAで迫る日本人の起源」 |date=23 December 2018 |type=Television production |language=ja |publisher=NHK |trans-title=The origin of Japanese people approaching with Yayoi DNA |location= |time= |people= |work=]}}</ref><ref>{{Cite journal |last1=Horai |first1=Satoshi |last2=Murayama |first2=Kumiko |date=1996 |title=mtDNA Polymorphism in East Asian Populations, with Special Reference to the Peopling of Japan |journal=] |location=Cambridge, Massachusetts |publisher=] |publication-date=1996 |volume=59 |issue=3 |pages=579–590 |pmc=1914908 |pmid=8751859}}</ref><ref>{{Cite journal |last1=Boer |first1=Elisabeth de |last2=Yang |first2=Melinda A. |last3=Kawagoe |first3=Aileen |last4=Barnes |first4=Gina L. |date=2020 |title=Japan considered from the hypothesis of farmer/language spread |journal=Evolutionary Human Sciences |language=en |volume=2 |pages=e13 |doi=10.1017/ehs.2020.7 |issn=2513-843X |pmc=10427481 |pmid=37588377 |doi-access=free}}</ref><ref name="Wang 2021 413–419" /> among other East Asian people. | |||
| ==== Autosomal DNA ==== | |||
| There is evidence of population admixture in ancient Korea. A 2022 study conducted by the ] found that an ancient Korea had considerable genetic diversity, including elevated levels of Jōmon ancestry.<ref name=":4" /> It was hypothesized that the Jōmon ancestry of ancient Koreans was lost over time, as they continually mixed with incoming populations from northern China, followed by a period of isolation during the Three Kingdoms period, resulting in the homogenous gene pool of modern Koreans.<ref name=":4" /><ref name=":1">{{cite web |title=1,700-year-old Korean genomes show genetic heterogeneity in Three Kingdoms period Gaya |url=https://www.sciencedaily.com/releases/2022/06/220621114705.htm |access-date=29 August 2022 |website=ScienceDaily |language=en}}</ref> | |||
| The majority of Japanese genetic ancestry is derived from sources related to other mainland Asian groups, mostly Koreans, while the other amount is derived from the local Jōmon hunter-gatherers.<ref name="Wang 2021 413–419" /> | |||
| According to a full genome analyses, the modern Japanese harbor a Northeast Asian (Amur River ancestry), an East Asian (Yellow River ancestry), and an indigenous Jōmon component. In addition to the indigenous Jōmon hunter-gatherers and the Yayoi period migrants, a new strand was hypothesized to have been introduced during the Yayoi-Kofun transition period that had strong cultural and political affinity with Korea and China.<ref>{{Cite web |last=Nikkei Science |date=23 June 2021 |title=渡来人、四国に多かった? ゲノムが明かす日本人ルーツ |trans-title=Were there many migrants in Shikoku? Japanese roots revealed by genome analysis |url=https://www.nikkei.com/article/DGXZQOUC18CCA0Y1A610C2000000/ |access-date=1 May 2022 |website=nikkei.com |language=ja}}</ref> However, this theory is mildly criticized in recent years (see ]). | |||
| Evidence for both Southern and Northern mtDNA and Y-DNA haplogroups has been observed in Koreans, similar to Japanese.<ref>{{cite journal | vauthors = Gakuhari T, Nakagome S, Rasmussen S, Allentoft ME, Sato T, Korneliussen T, Chuinneagáin BN, Matsumae H, Koganebuchi K, Schmidt R, Mizushima S, Kondo O, Shigehara N, Yoneda M, Kimura R, Ishida H, Masuyama T, Yamada Y, Tajima A, Shibata H, Toyoda A, Tsurumoto T, Wakebe T, Shitara H, Hanihara T, Willerslev E, Sikora M, Oota H | display-authors = 6 | title = Ancient Jomon genome sequence analysis sheds light on migration patterns of early East Asian populations | journal = Communications Biology | volume = 3 | issue = 1 | pages = 437 | date = August 2020 | pmid = 32843717 | pmc = 7447786 | doi = 10.1038/s42003-020-01162-2 }}</ref> | |||
| ==== Paternal lineages ==== | ==== Paternal lineages ==== | ||
| A comprehensive study of worldwide Y-DNA diversity (Underhill ''et al.'' 2000) included a sample of 23 males from Japan, of whom 35% belonged to ], 26% belonged to ], 22% belonged to ], 13% belonged to ] and ], and 4.3% belonged to ].<ref>{{cite journal |display-authors=6 |vauthors=Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonné-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi SQ, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza LL, Oefner PJ |date=November 2000 |title=Y chromosome sequence variation and the history of human populations |journal=Nature Genetics |volume=26 |issue=3 |pages=358–61 |doi=10.1038/81685 |pmid=11062480 |s2cid=12893406}}</ref> Poznik ''et al.'' (2016) reported the haplogroups of a sample of Japanese men from Tokyo:<ref>The JPT sample is considered "as generally representative of the majority population in Japan". See {{Cite web |title=Japanese in Tokyo, Japan – Population Description |url=https://catalog.coriell.org/0/Sections/Collections/NHGRI/Japanese.aspx?PgId=359 |publisher=Coriell Institute for Medical Research |location=Camden, ] |vauthors=Matsuda I}}</ref> 36% belonged to ], 32% had ], 18% carried ], 7.1% carried ], 3.6% belonged to ], and 3.6% carried ].<ref>{{Cite journal |display-authors=6 |vauthors=Poznik GD, Xue Y, Mendez FL, Willems TF, Massaia A, Wilson Sayres MA, Ayub Q, McCarthy SA, Narechania A, Kashin S, Chen Y, Banerjee R, Rodriguez-Flores JL, Cerezo M, Shao H, Gymrek M, Malhotra A, Louzada S, Desalle R, Ritchie GR, Cerveira E, Fitzgerald TW, Garrison E, Marcketta A, Mittelman D, Romanovitch M, Zhang C, Zheng-Bradley X, Abecasis GR, McCarroll SA, Flicek P, Underhill PA, Coin L, Zerbino DR, Yang F, Lee C, Clarke L, Auton A, Erlich Y, Handsaker RE, Bustamante CD, Tyler-Smith C |date=June 2016 |title=Punctuated bursts in human male demography inferred from 1,244 worldwide Y-chromosome sequences |journal=Nature Genetics |volume=48 |issue=6 |pages=593–9 |doi=10.1038/ng.3559 |pmc=4884158 |pmid=27111036 |hdl=11858/00-001M-0000-002A-F024-C}}</ref> | |||
| Jin Han-jun et al. (2003)<!--The article's abstract wrote this person's name as "Han-Jun Jin." Jin Han-jun is this person's name in the Korean name ordering.--> said that the distribution of ] shows that Koreans have a complex origin that results from genetic contributions from ], most of which are from a southern-to-northern region in the continent, and genetic contributions from the northern Asian settlement.<!--This information is in the last sentence of the article's abstract.--><ref name="Jin_2003">{{Cite journal |vauthors=Jin HJ, Kwak KD, Hammer MF, Nakahori Y, Shinka T, Lee JW, Jin F, Jia X, Tyler-Smith C, Kim W |date=December 2003 |title=Y-chromosomal DNA haplogroups and their implications for the dual origins of the Koreans |journal=Human Genetics |volume=114 |issue=1 |pages=27–35 |doi=10.1007/s00439-003-1019-0 |pmid=14505036 |s2cid=1396796}}</ref> | |||
| The most common Y-DNA haplogroup among present-day Koreans is ] (O2, formerly O3), which is a very ancient (formed 33,943 | |||
| years before present, TMRCA 30,365 years before present according to Karmin ''et al.'' 2022<ref name = "Karmin2022">Monika Karmin, Rodrigo Flores, Lauri Saag, Georgi Hudjashov, Nicolas Brucato, Chelzie Crenna-Darusallam, Maximilian Larena, Phillip L. Endicott, Mattias Jakobsson, J. Stephen Lansing, Herawati Sudoyo, Matthew Leavesley, Mait Metspalu, François-Xavier Ricaut, and Murray P. Cox (2022), "Episodes of Diversification and Isolation in Island Southeast Asian and Near Oceanian Male Lineages." ''Mol. Biol. Evol.'' 39(3): msac045 doi:10.1093/molbev/msac045</ref>) and frequently observed Y-DNA haplogroup among East and Southeast Asians in general.<ref name="Shi2005">{{Cite journal |vauthors=Shi H, Dong YL, Wen B, Xiao CJ, Underhill PA, Shen PD, Chakraborty R, Jin L, Su B |date=September 2005 |title=Y-chromosome evidence of southern origin of the East Asian-specific haplogroup O3-M122 |journal=American Journal of Human Genetics |volume=77 |issue=3 |pages=408–19 |doi=10.1086/444436 |pmc=1226206 |pmid=16080116}}</ref><ref name="Wen2004"/><ref name="Karmin2015">{{Cite journal |vauthors=Karmin M, Saag L, Vicente M, Wilson Sayres MA, Järve M, Talas UG, etal |date=April 2015 |title=A recent bottleneck of Y chromosome diversity coincides with a global change in culture |journal=Genome Research |volume=25 |issue=4 |pages=459–66 |doi=10.1101/gr.186684.114 |pmc=4381518 |pmid=25770088}}</ref> Haplogroup O2-M122 has been found in approximately 41% of sampled Korean males, including 23/79 = 29.1% ],<ref name="Katoh2005">{{Cite journal |display-authors=6 |vauthors=Katoh T, Munkhbat B, Tounai K, Mano S, Ando H, Oyungerel G, Chae GT, Han H, Jia GJ, Tokunaga K, Munkhtuvshin N, Tamiya G, Inoko H |date=February 2005 |title=Genetic features of Mongolian ethnic groups revealed by Y-chromosomal analysis |journal=Gene |volume=346 |pages=63–70 |doi=10.1016/j.gene.2004.10.023 |pmid=15716011}}</ref> 29/85 = 34.1% Korean (collected in Seoul by the Catholic University of Korea),<ref name="Katoh2005"/> 16/45 = 35.6% Korean,<ref name="Wells2001">{{Cite journal |display-authors=6 |vauthors=Wells RS, Yuldasheva N, Ruzibakiev R, Underhill PA, Evseeva I, Blue-Smith J, Jin L, Su B, Pitchappan R, Shanmugalakshmi S, Balakrishnan K, Read M, Pearson NM, Zerjal T, Webster MT, Zholoshvili I, Jamarjashvili E, Gambarov S, Nikbin B, Dostiev A, Aknazarov O, Zalloua P, Tsoy I, Kitaev M, Mirrakhimov M, Chariev A, Bodmer WF |date=August 2001 |title=The Eurasian heartland: a continental perspective on Y-chromosome diversity |journal=Proceedings of the National Academy of Sciences of the United States of America |volume=98 |issue=18 |pages=10244–10249 |bibcode=2001PNAS...9810244W |doi=10.1073/pnas.171305098 |pmc=56946 |pmid=11526236 |doi-access=free}}</ref> 109/300 = 36.3% unrelated Korean males obtained from the National Biobank of Korea,<ref name="Park2013"/> 31/84 = 36.9% Gyeongsang,<ref name="Kim2011" /> 24/63 = 38.1% Gangwon,<ref name="Kim2011" /> 17/43 = 39.5% Korean (Korea),<ref name="Xue2006" /> 10/25 = 40.0% ],<ref name="Xue2006" /> 30/75 = 40.0% Korea,<ref name="Hammer2006" /> 55/133 = 41.4% Daejeon,<ref name="Park2012">{{Cite journal |vauthors=Park MJ, Lee HY, Yang WI, Shin KJ |date=July 2012 |title=Understanding the Y chromosome variation in Korea—relevance of combined haplogroup and haplotype analyses |journal=International Journal of Legal Medicine |volume=126 |issue=4 |pages=589–99 |doi=10.1007/s00414-012-0703-9 |pmid=22569803 |s2cid=27644576}}</ref> 23/55 = 41.8% Korean from the KoVariome database,<ref name="Kim2020">{{Cite journal |vauthors=Kim J, Jeon S, Choi JP, Blazyte A, Jeon Y, Kim JI, Ohashi J, Tokunaga K, Sugano S, Fucharoen S, Al-Mulla F, Bhak J |date=May 2020 |title=The Origin and Composition of Korean Ethnicity Analyzed by Ancient and Present-Day Genome Sequences |journal=Genome Biology and Evolution |volume=12 |issue=5 |pages=553–565 |doi=10.1093/gbe/evaa062 |pmc=7250502 |pmid=32219389}}</ref> 65/154 = 42.2% Korean,<ref name="Hong2006">Hong, S., Jin, H., Kwak, K., & Kim, W. (2006). "Y-chromosome Haplogroup O3-M122 Variation in East Asia and Its Implication for the Peopling of Korea." ''Genes & Genomics'', 28, 1–8.</ref><ref name="Jin2009"/> 242/573 = 42.2% Seoul,<ref name="Park2012"/> 39/90 = 43.3% Jeolla,<ref name="Kim2011" /> 38/87 = 43.7% Jeju,<ref name="Kim2011" /> 102/216 = 47.2% Seoul & Daejeon,<ref name="Kim2007">{{Cite journal |vauthors=Kim W, Yoo TK, Kim SJ, Shin DJ, Tyler-Smith C, Jin HJ, Kwak KD, Kim ET, Bae YS |date=January 2007 |title=Lack of association between Y-chromosomal haplogroups and prostate cancer in the Korean population |journal=PLOS ONE |volume=2 |issue=1 |pages=e172 |bibcode=2007PLoSO...2..172K |doi=10.1371/journal.pone.0000172 |pmc=1766463 |pmid=17245448 |doi-access=free}} {{open access}}</ref> 36/72 = 50.0% Chungcheong,<ref name="Kim2011" /> 56/110 = 50.9% Seoul-Gyeonggi.<ref name="Kim2011" /> | |||
| The second most common Y-DNA haplogroup among present-day Koreans is ] (O1b2, formerly O2b), a clade that probably has spread mainly from somewhere in the Korean Peninsula or its vicinity.<ref name="Kim2011" /><ref name="Balaresque2015">{{Cite journal |vauthors=Balaresque P, Poulet N, Cussat-Blanc S, Gerard P, Quintana-Murci L, Heyer E, Jobling MA |date=October 2015 |title=Y-chromosome descent clusters and male differential reproductive success: young lineage expansions dominate Asian pastoral nomadic populations |journal=European Journal of Human Genetics |volume=23 |issue=10 |pages=1413–22 |doi=10.1038/ejhg.2014.285 |pmc=4430317 |pmid=25585703}}</ref> Haplogroup O1b2-M176 has been found in approximately 31% of sampled Korean males (including 31/154 = 20.1% Korean,<ref name="Jin2009"/> 58/216 = 26.9% ] & ],<ref name="Kim2007"/> 12/43 = 27.9% Koreans in Korea,<ref name="Xue2006" /> 7/25 = 28.0% ],<ref name="Xue2006" /> 31/110 = 28.2% ]-],<ref name="Kim2011" /> 25/84 = 29.8% ],<ref name="Kim2011" /> 22/72 = 30.6% ],<ref name="Kim2011" /> 28/90 = 31.1% ],<ref name="Kim2011" /> 14/45 = 31.1% O-M175(xO1a-M119, O2a1-M95, O3-M122) Korean,<ref name="Wells2001"/> 181/573 = 31.6% Seoul,<ref name="Park2012"/> 28/87 = 32.2% ],<ref name="Kim2011" /> 28/85 = 32.9% Korean (collected in Seoul by the Catholic University of Korea),<ref name="Katoh2005"/> 45/133 = 33.8% ],<ref name="Park2012"/> 28/75 = 37.3% Korea,<ref name="Hammer2006" /> 113/300 = 37.7% unrelated Korean males obtained from the National Biobank of Korea,<ref name="Park2013"/> 25/63 = 39.7% ],<ref name="Kim2011" /> 38/79 = 48.1% ]<ref name="Katoh2005"/>).<ref name="Xue2006" /><ref name="Hammer_2006">{{Cite journal |vauthors=Hammer MF, Karafet TM, Park H, Omoto K, Harihara S, Stoneking M, Horai S |date=2006 |title=Dual origins of the Japanese: common ground for hunter-gatherer and farmer Y chromosomes |journal=Journal of Human Genetics |volume=51 |issue=1 |pages=47–58 |doi=10.1007/s10038-005-0322-0 |pmid=16328082 |doi-access=free}}</ref><ref name="Jin_2003"/><ref name="Kim2007"/><ref name="Shin2001">{{Cite journal |vauthors=Shin DJ, Jin HJ, Kwak KD, Choi JW, Han MS, Kang PW, Choi SK, Kim W |date=October 2001 |title=Y-chromosome multiplexes and their potential for the DNA profiling of Koreans |journal=International Journal of Legal Medicine |volume=115 |issue=2 |pages=109–17 |doi=10.1007/s004140100248 |pmid=11724428 |s2cid=27739773}}</ref> | |||
| Korean males also exhibit a moderate frequency (approximately 15%) of ]. About 2% of Korean males belong to ] (0/216 = 0.0% DE-YAP,<ref name="Kim2007"/> 3/300 = 1.0% DE-M145,<ref name="Park2013">{{Cite journal |vauthors=Park MJ, Lee HY, Kim NY, Lee EY, Yang WI, Shin KJ |date=January 2013 |title=Y-SNP miniplexes for East Asian Y-chromosomal haplogroup determination in degraded DNA |journal=Forensic Science International. Genetics |volume=7 |issue=1 |pages=75–81 |doi=10.1016/j.fsigen.2012.06.014 |pmid=22818129}}</ref> 1/68 = 1.5% DE-YAP(xE-SRY4064),<ref name="Xue2006" /> 8/506 = 1.6% ],<ref name="Kim2011" /> 3/154 = 1.9% DE, 18/706 = 2.55% D-M174,<ref name="Kwon2015">{{Cite journal |vauthors=Kwon SY, Lee HY, Lee EY, Yang WI, Shin KJ |date=November 2015 |title=Confirmation of Y haplogroup tree topologies with newly suggested Y-SNPs for the C2, O2b and O3a subhaplogroups |journal=Forensic Science International. Genetics |volume=19 |pages=42–46 |doi=10.1016/j.fsigen.2015.06.003 |pmid=26103100}}</ref> 5/164 = 3.0% D-M174,<ref name="Katoh2005"/> 1/75 D1b*-P37.1(xD1b1-M116.1) + 2/75 D1b1a-M125(xD1b1a1-P42) = 3/75 = 4.0% D1b-P37.1,<ref name="Hammer_2006"/> 3/45 = 6.7% D-M174<ref>{{cite journal |display-authors=6 |vauthors=Wells RS, Yuldasheva N, Ruzibakiev R, Underhill PA, Evseeva I, Blue-Smith J, Jin L, Su B, Pitchappan R, Shanmugalakshmi S, Balakrishnan K, Read M, Pearson NM, Zerjal T, Webster MT, Zholoshvili I, Jamarjashvili E, Gambarov S, Nikbin B, Dostiev A, Aknazarov O, Zalloua P, Tsoy I, Kitaev M, Mirrakhimov M, Chariev A, Bodmer WF |date=August 2001 |title=The Eurasian heartland: a continental perspective on Y-chromosome diversity |journal=Proceedings of the National Academy of Sciences of the United States of America |volume=98 |issue=18 |pages=10244–9 |bibcode=2001PNAS...9810244W |doi=10.1073/pnas.171305098 |pmc=56946 |pmid=11526236 |doi-access=free}}</ref>). The D1b-M55 subclade has been found with maximal frequency in a small sample (n=16) of the ] of Japan, and is generally frequent throughout the Japanese Archipelago.<ref name="Tajima2004">{{Cite journal |vauthors=Tajima A, Hayami M, Tokunaga K, Juji T, Matsuo M, Marzuki S, Omoto K, Horai S |year=2004 |title=Genetic origins of the Ainu inferred from combined DNA analyses of maternal and paternal lineages |journal=Journal of Human Genetics |volume=49 |issue=4 |pages=187–93 |doi=10.1007/s10038-004-0131-x |pmid=14997363 |doi-access=free}}</ref> Other haplogroups that have been found less commonly in samples of Korean males are Y-DNA ] (approx. 4%), ] (approx. 3%), ](xM176) (approx. 2%), ] and ] (approx. 2% total), J, Y*(xA, C, DE, J, K), L, C-RPS4Y(xM105, M38, M217), and C-M105.<ref name="Kim2011" /><ref name="Xue2006" /> | |||
| He Miao<!--The article wrote the name "Miao He". "He Miao" is the person's name in the Chinese name order.--> et al. (2009) created an artificial combination of equal parts of the ] of the ] samples of Han Chinese in Beijing and Japanese in Tokyo. The study said that this artificial combination resembled five populations which included Koreans in South Korea and ].<!--This information is in the second sentence of the third paragraph of the right column of page 5 of 8 of the PDF document. That sentence uses the acronyms "CHB" and "JPT". These two acronyms are defined in the second sentence of the "Introduction" section of page 1 of 8 of the PDF document as "Han Chinese in Beijing, China" and "Japanese in Tokyo, Japan," respectively. That sentence uses the phrase "Chinese Koreans" which could ambiguously mean either Koreans in China or Chinese in Korea. Figure 3 Map "A" on page 6 of 8 of the PDF document shows a dot for "Chinese Korean" in China north of North Korea, so the study must be using the term "Chinese Koreans" to mean an analyzed regional population of Koreans in China rather than Chinese in Korea. The same sentence uses the term "Koreans," and Figure 3 Map "A" on page 6 of 8 of the PDF document shows the dot for "Korean" in South Korea, so the study, in that sentence, is using the singular term "Korean" to mean an analyzed regional population of Koreans in South Korea.--><ref>{{Cite journal |vauthors=He M, Gitschier J, Zerjal T, de Knijff P, Tyler-Smith C, Xue Y |year=2009 |title=Geographical affinities of the HapMap samples |journal=PLOS ONE |volume=4 |issue=3 |pages=e4684 |bibcode=2009PLoSO...4.4684H |doi=10.1371/journal.pone.0004684 |pmc=2649532 |pmid=19259268 |doi-access=free}}</ref> | |||
| ==== Maternal lineages ==== | ==== Maternal lineages ==== | ||
| According to an analysis of the ]'s sample of Japanese collected in the Tokyo metropolitan area, the mtDNA haplogroups found among modern Japanese include ] (35.6%), ] (13.6%), ] (10.2%), ] (10.2%), ] (8.5%), ] (7.6%), ] (6.8%), ] (3.4%), ] (2.5%), and ] (1.7%).<ref>{{Cite journal |vauthors=Zheng HX, Yan S, Qin ZD, Wang Y, Tan JZ, Li H, Jin L |year=2011 |title=Major population expansion of East Asians began before neolithic time: evidence of mtDNA genomes |journal=PLOS ONE |volume=6 |issue=10 |pages=e25835 |bibcode=2011PLoSO...625835Z |doi=10.1371/journal.pone.0025835 |pmc=3188578 |pmid=21998705 |doi-access=free}}</ref> | |||
| Studies of Korean ] lineages have shown that there is a high frequency of ], ranging from approximately 23% among ethnic Koreans in ], ]<ref name="Kong_2003">{{Cite journal |vauthors=Kong QP, Yao YG, Liu M, Shen SP, Chen C, Zhu CL, Palanichamy MG, Zhang YP |date=October 2003 |title=Mitochondrial DNA sequence polymorphisms of five ethnic populations from northern China |journal=Human Genetics |volume=113 |issue=5 |pages=391–405 |doi=10.1007/s00439-003-1004-7 |pmid=12938036 |s2cid=6370358}}</ref> to approximately 32% among Koreans from South Korea.<ref name="Jin_2009">{{Cite journal |vauthors=Jin HJ, Tyler-Smith C, Kim W |year=2009 |title=The peopling of Korea revealed by analyses of mitochondrial DNA and Y-chromosomal markers |journal=PLOS ONE |volume=4 |issue=1 |pages=e4210 |bibcode=2009PLoSO...4.4210J |doi=10.1371/journal.pone.0004210 |pmc=2615218 |pmid=19148289 |doi-access=free}} {{open access}}</ref><ref name="Derenko2007">{{Cite journal |display-authors=6 |vauthors=Derenko M, Malyarchuk B, Grzybowski T, Denisova G, Dambueva I, Perkova M, Dorzhu C, Luzina F, Lee HK, Vanecek T, Villems R, Zakharov I |date=November 2007 |title=Phylogeographic analysis of mitochondrial DNA in northern Asian populations |journal=American Journal of Human Genetics |volume=81 |issue=5 |pages=1025–41 |doi=10.1086/522933 |pmc=2265662 |pmid=17924343}}</ref> | |||
| Haplogroup D4 is the modal mtDNA haplogroup among northern East Asians in general, with a peak frequency among Japanese and Ryukyuans in Japan. ], which occurs very frequently in many populations of Southeast Asia, Polynesia, and the Americas, is found in approximately 10% to 20% of Koreans.<ref name="Kong_2003"/><ref name="Derenko2007"/> ] has been detected in approximately 7% to 15% (7/48 ethnic Koreans from ], Inner Mongolia) of Koreans.<ref name="Jin_2009"/><ref name="Kong_2003"/><ref name="Derenko2007"/> Haplogroup A is the most common mtDNA haplogroup among the ], ], ], and many ] ethnic groups of North and Central America. | |||
| === Korean people === | |||
| The other half of the Korean mtDNA pool consists of an assortment of various haplogroups, each found with relatively low frequency, such as ], ], ], ], ], ], ], ], ], ], ], ], and ]. | |||
| {{Main|Koreans}}Korean populations in modern Korea can be traced to many origins from the people of the ] to the ].<ref>{{cite book |last1=Preucel |first1=Robert |title=Contemporary Archaeology in Theory: The New Pragmatism |last2=Mrozowski |first2=Stephen |last3=Nelson |first3=Sarah |date=2010 |publisher=Wiley-Blackwell |edition=2nd |pages=218–221}}</ref> In modern times, Koreans are related to other populations found in Northeast Asia, however according to recent studies, ancient Koreans included populations related to the ],<ref>{{Cite web |last=水野 |first=文月 |date=15 October 2024 |title=弥生時代人の古代ゲノム解析から渡来人のルーツを探る |url=https://www.s.u-tokyo.ac.jp/ja/press/10527/ |access-date= |website=東京大学 大学院理学系研究科・理学部 |language=ja}}</ref> ],<ref name=":42" /> ] influx<ref name="Siska2017">{{cite journal |last1=Siska |first1=Veronika |last2=Jones |first2=Eppie |last3=Jeon |first3=Sungwon |display-authors=etal |year=2017 |title=Genome-wide data from two early Neolithic East Asian individuals dating to 7700 years ago |journal=Science Advances |volume=3 |issue=2 |pages=e1601877 |doi=10.1126/sciadv.1601877 |pmid=28164156 |pmc=5287702 |bibcode=2017SciA....3E1877S |hdl-access=free |hdl=2262/90843}}</ref> and etc.] | |||
| Hwan Young Lee ''et al.'' (2006) studied a sample of 694 Koreans and found the following mtDNA distribution: 32.56% ], 14.84% ], 9.56% ], 8.79% ], 8.36% ], 8.21% ], 7.78% ], 4.76% ], 1.01% M8a, 1.01% ], 1.87% ], 1.73% ], 0.72% ], 0.29% ], 0.14% ], 0.14% ], and 0.14% ].<ref name="Lee2006">{{Cite journal |vauthors=Lee HY, Yoo JE, Park MJ, Chung U, Kim CY, Shin KJ |date=November 2006 |title=East Asian mtDNA haplogroup determination in Koreans: haplogroup-level coding region SNP analysis and subhaplogroup-level control region sequence analysis |journal=Electrophoresis |volume=27 |issue=22 |pages=4408–4418 |doi=10.1002/elps.200600151 |pmid=17058303 |s2cid=28252456}}</ref> | |||
| Derenko ''et al.'' (2007) examined a sample of 103 Koreans from South Korea and found that they belonged to the following mtDNA haplogroups: 39.8% D, 20.4% B, 9.7% M7, 6.8% A, 6.8% G, 4.9% F1, 4.9% M8a2, 2.9% N9a, 1.9% M9a, 1.0% C, and 1.0% Y.<ref name="Derenko2007"/> | |||
| Similar studies have reported varying frequencies of the above haplogroups across various regions of Korea, based on large sample sizes.<ref name="Hong2014">{{Cite journal |vauthors=Hong SB, Kim KC, Kim W |year=2014 |title=Mitochondrial DNA haplogroups and homogeneity in the Korean population |journal=Genes & Genomics |volume=36 |issue=5 |pages=583–590 |doi=10.1007/s13258-014-0194-9 |s2cid=16827252}}</ref><ref name="Jeon2020">{{cite journal | vauthors = Jeon S, Bhak Y, Choi Y, Jeon Y, Kim S, Jang J, Jang J, Blazyte A, Kim C, Kim Y, Shim J, Kim N, Kim YJ, Park SG, Kim J, Cho YS, Park Y, Kim HM, Kim BC, Park NH, Shin ES, Kim BC, Bolser D, Manica A, Edwards JS, Church G, Lee S, Bhak J | display-authors = 6 | title = Korean Genome Project: 1094 Korean personal genomes with clinical information | journal = Science Advances | volume = 6 | issue = 22 | pages = eaaz7835 | date = May 2020 | pmid = 32766443 | pmc = 7385432 | doi = 10.1126/sciadv.aaz7835 | bibcode = 2020SciA....6.7835J | doi-access = free }}</ref> The individuals sampled for the Korean Genome Project are mostly from the ] metropolitan region.<ref name="Jeon2020"/> | |||
| ==== Autosomal DNA ==== | ==== Autosomal DNA ==== | ||
| Ancient genome comparisons revealed that the genetic makeup of Koreans can be best described as an admixture between Northeast Asian hunter-gatherers and an influx of rice-farming Southeast Asian agriculturalists from the Yangtze river valley.<ref name="Jin" /> This is supported by archaeological, historical and linguistic evidence, which suggest that the direct ancestors of Koreans were proto-Koreans who inhabited the northeastern region of China and the Korean Peninsula during the Neolithic (8,000–1,000 BC) and Bronze (1,500–400 BC) Ages.<ref name="Kim2011" /> | |||
| Jin Han-jun et al. (1999)<!--The study wrote the first author's name as "Han Jun Jin." Jin Han-jun is the first author's name in the Korean name ordering.--> said that based on genetic studies of classic ]s of ] and ], Koreans tend to be closely genetically related to Mongols among East Asians, which is supported by the following studies: Goedde et al. (1987); Saha & Tay (1992); Hong et al. (1993); and Nei & Roychoudhury (1993).<!--The preceding information is in the first sentence of the first paragraph of the left column of page 2/6 of the PDF document (page 393).--> The study said that the ] 9‐] deletion frequency in the ] '']/]<sup>]</sup>'' region of Mongols (5.1%) is lower than that of Chinese (14.2%), Japanese (14.3%) and Koreans (15.5%). The study said that these 9‐bp deletion frequencies suggest that Koreans are closely related to Japanese and Chinese and that Koreans are not so closely related to Mongols.<!--The preceding information is in the first and second sentences of the second paragraph of the left column of page 5/6 of the PDF document (page 396). The frequency numbers for Chinese, Japanese, and Koreans are in the "9-bp deletion" column of Table 1 of page 4/6 of the PDF document (page 395).--> The study said that the ] in the 9-bp deletion frequencies among Chinese (14.2%), Japanese (14.3%), and Koreans (15.5%), only spanning from a low of 14.2% for Chinese to a high of 15.5% for Koreans, indicates that very few ] are differentiated in these three populations.<!--The preceding information is in the second-to-last sentence of the first paragraph of the left column of page 5/6 of the PDF document (page 396). The frequency numbers for Chinese, Japanese, and Koreans are in the "9-bp deletion" column of Table 1 of page 4/6 of the PDF document (page 395).--> The study said that the 9‐bp deletion frequencies for Vietnamese (23.2%) and ] (25.0%), which are the two populations constituting Mongoloid Southeast Asians in the study, are relatively high frequencies when compared to the 9-bp deletion frequencies for Mongols(5.1%), Chinese (14.2%), Japanese (14.3%) and Koreans (15.5%), which are the four populations constituting Northeast Asians in the study. The study said that these 9-bp deletion frequencies are consistent with earlier surveys which showed that 9-bp deletion frequencies increase going from Japan to mainland Asia to the ], which is supported by the following studies: Horai et al. (1987); Hertzberg et al. (1989); Stoneking & Wilson (1989); Horai (1991); Ballinger et al. (1992); Hanihara et al. (1992); and Chen et al. (1995).<!--The preceding information is in the second, third and fourth sentences of the second paragraph of the right column of page 4/6 of the PDF document (page 395).--> The study said that ], from Cavalli-Sforza & Bodmer (1971), which is based on the ] frequencies of the intergenic ''COII/tRNA<sup>Lys</sup>'' region, showed that Koreans are more genetically related to Japanese than Koreans are genetically related to the other East Asian populations which were surveyed.<!--The preceding information is in the first sentence of the third paragraph of the left column of page 5/6 of the PDF document (page 396). The caption in Table 2, which is in page 5/6 of the PDF document (page 396), says, "Cavalli-Sforza's cord genetic distance (4D) in the Table body was based on allele frequency in the intergenic ''COII/tRNA<sup>Lys</sup>'' region." The caption in Table 2 misspelled the word "chord" as "cord."--> The Cavalli-Sforza's chord genetic distance (4D) between Koreans and other East Asian populations in the study, from least to greatest, are as follows: Korean to Japanese (0.0019), Korean to Chinese (0.0141), Korean to Vietnamese (0.0265), Korean to Indonesian (0.0316) and Korean to Mongols (0.0403).<!--The preceding information is in Table 2 of page 5/6 of the PDF document (page 396).--> The study said that the close genetic affinity between present-day Koreans and Japanese is expected due to the ] migration from China and the Korean Peninsula to Japan which began about 2,300 years ago, a migration that is supported by the following studies: Chard (1974); Hanihara (1991); Hammer & Horai (1995); Horai et al. (1996); Omoto & Saitou (1997).<!--The preceding information is in the second and third sentences of the third paragraph of the left column of page 5/6 of the PDF document (page 396).--> The study said that Horai et al. (1996) detected ] ] variation which supports the idea that a large amount of ] came into Japan from immigrants from the Korean Peninsula after the ].<!--The preceding information is in the second-to-last sentence of the third paragraph of the left column of page 5/6 of the PDF document (page 396).--><ref name="JinHanjun1999">{{Cite journal |vauthors=Jin HJ, Choi JW, Shin DJ, Kim JM, Kim W |year=1999 |title=Distribution of length variation of the mtDNA 9‐bp motif in the intergenic COII/tRNALys region in East Asian populations |journal=Korean Journal of Biological Sciences |volume=3 |issue=4 |pages=393–397 |doi=10.1080/12265071.1999.9647513 |doi-access=free}}</ref> | |||
| There is evidence for considerable genetic diversity,<ref name=":42" /> including elevated levels of Jōmon ancestry among early southern Koreans.<ref>{{harvnb|Wang|Wang|2022|ps=: "The genetic legacy of Jomon was not restricted to Japan but was also found in Neolithic Korea."}}</ref> It was hypothesized that the Jōmon ancestry of ancient Koreans was lost over time, as they continually mixed with incoming populations from northern China,<ref>{{harvnb|Wang|Wang|2022|ps=: "Taking no account of this Bronze Age sample, the possible scenario was that the indigenous Jomon-related ancestry was largely replaced through admixture with the incoming northern China populations and disappeared in the later centuries, creating relatively homogeneous present-day Koreans."}}</ref> followed by a period of isolation during the Three Kingdoms period, resulting in the homogenous gene pool of modern Koreans.<ref>{{harvnb|Wang|Wang|2022|ps=: "Present-day Koreans are a highly homogeneous population without apparent genetic substructure on a whole-genome scale and with close genetic relationships with Japanese and northern Han Chinese. The genetic variation of the Y chromosome and mitochondrial DNA show that Koreans contain lineages typical of both Southeast and Northeast Asian populations, suggesting that Korea was populated in multiple waves"}}</ref><ref name=":42" /> | |||
| Wook<!--The article writes this person's name as "Wook Kim." "Kim Wook" is this person's name in the Korean name order.--> et al. (2000) said that Chu et al. (1998) found that ] which was based on 30 ]s indicated that Korean people were closely related to Chinese people from ] and ], but Kim Wook et al. (2000) found that the high incidence of the DXYS156Y-null variant in northeast Chinese implied that it is possible to exclude these northeastern Chinese populations from being sources which are significant in Korean people.<!--The preceding information is in the sixth sentence which starts with "Interestingly" and the eighth sentence which starts with "The relatively high" of the third paragraph of the left column of page 8/9 of the PDF document (page 82).--> The phylogenetic analysis done by Wook et al. (2000) indicated that Japanese people are genetically closer to Korean people than Japanese people are genetically related to any of the following peoples: Mongolians, Chinese, Vietnamese, Indonesians, Filipinos, and Thais. The study said that mainland Japanese having Koreans as their closest genetic population is consistent with the following previous studies: Hammer and Horai (1995); Horai et al. (1996); and Kim et al. (1998).<!--The preceding information is in the ninth sentence which starts with "The phylogeny also indicated" and the tenth sentence which starts with "This result is" which are both in the third paragraph of the left column of page 8/9 of the PDF document (page 82). The "phylogeny" referenced in the ninth sentence refers to the first sentence of the same paragraph which said, "Phylogenetic analysis also reflected a considerable difference between Southeast and Northeast Asian populations (Fig. 3)." The groups indicated by the phrase, "other Asian populations surveyed," can be seen in Table 1 or Table 2 or Figure 3 or the "DNA samples" section of the "Subjects and methods" section to be Mongolians, Chinese, Vietnamese, Indonesians, Filipinos, and Thais.--> The study found that Koreans are more genetically ] than the Japanese, and the study said that this might be due to different sizes of the ] and ].<!--The preceding information is in the first sentence of the first paragraph of the right column of page 8/9 of the PDF document (page 82).--> The study said that the moderate ] ] diversity value for Koreans might be the result of migrations from East Asia that had a homogenizing influence.<!--The preceding information is in the last sentence of the second paragraph of page 7/9 of the PDF document (page 81). The "Korean variability (0.482)" referenced in that sentence is the same thing as the "diversity values (h)" referenced in the third sentence of the fourth paragraph of the left column of page 6/9 of the PDF document (page 80). The variable "h" is indicated to be "mean haplotype diversity" in the second sentence of the fourth paragraph of the right column of page 4/9 of the PDF document (page 78). The fourth sentence clarifies that the "dual patterns of the haplotype distribution" referenced in the last sentence refer to Koreans.--><ref name="Kim_2000">{{Cite journal |vauthors=Kim W, Shin DJ, Harihara S, Kim YJ |date=2000 |title=Y chromosomal DNA variation in east Asian populations and its potential for inferring the peopling of Korea |journal=Journal of Human Genetics |volume=45 |issue=2 |pages=76–83 |doi=10.1007/s100380050015 |pmid=10721667 |doi-access=free}}</ref> | |||
| A 2022 study was unable to detect significant Jōmon ancestry in modern Koreans, however by using different proxies of ancestry, a Jōmon contribution of 3.1–4.4% was found for present-day Ulsan Koreans. Nevertheless, the authors suggested that the model that yielded this result is not the most reliable.<ref>{{cite journal |last1=Lee |first1=Don-Nyeong |last2=Jeon |first2=Chae Lin |last3=Kang |first3=Jiwon |last4=Burri |first4=Marta |last5=Krause |first5=Johannes |last6=Woo |first6=Eun Jin |last7=Jeong |first7=Choongwon |date=December 2022 |title=Genomic detection of a secondary family burial in a single jar coffin in early Medieval Korea |journal=American Journal of Biological Anthropology |language=en |volume=179 |issue=4 |pages=585–597 |doi=10.1002/ajpa.24650 |issn=2692-7691 |doi-access=free|pmc=9827920}} "In both ancient and present-day Koreans, we do not detect a statistically significant contribution from the Jomon hunter-gatherer gene pool of the Japanese archipelago (Table S7A), although previous studies report occasional presence of the Jomon ancestry contribution from Neolithic to the early Medieval period (Gelabert et al., 2022; Robbeets et al., 2021). When we replace the genetic northern proxy from WLR_BA to Middle Neolithic individuals from the Miaogizou site in Inner Mongolia (“Miaozigou_MN”), we detect a small but significant amount of Jomon contribution in the Gunsan individuals and present-day Ulsan Koreans (3.1%–4.4%; Table S7B). We believe that WLR_BA provides a more suitable model for ancient and present-day Koreans given its geographical and temporal proximity to them. The remaining well-fitting source pairs provide qualitatively similar results (Table S8)."</ref> | |||
| Evidence for both Southern and Northern mtDNA and Y-DNA haplogroups has been observed in Koreans, similar to Japanese.<ref name="auto3" /> | |||
| Kim Jong-jin<!--The study wrote Kim Jong-jin's name as "Jong-Jin Kim".--> et al. (2005) did a study about the genetic relationships among East Asians based on ], particularly focusing on how close Chinese, Japanese and Koreans are genetically related to each other.<!--This information is in the first sentence of the article's abstract on page 511 which is page 1/9 of the PDF document. The phrase "particularly focusing on how... are related" is a rewording of the source text's phrase "with a specific focus on the relationships among".--> Most Koreans were hard to distinguish from Japanese,<!--This information is in the second-to-last sentence of the article's abstract on page 511 which is page 1/9 of the PDF document.--> and the study was not able to clearly distinguish Koreans and Japanese.<!--This information is in the second sentence of the third paragraph of the right column of page 518 which is page 8/9 of the PDF document.--> Koreans and Japanese clustered together in the ] and the best ] tree.<!--This information is in the third-to-last sentence of the article's abstract on page 511 which is page 1/9 of the PDF document.--> The study said that "''ommon ancestry and/or extensive gene flow''" historically between Koreans and Japanese appears to be "''likely''" and results in a lot of difficulty finding population-specific alleles that could assist in differentiating Koreans and Japanese.<!--This information is in the fourth sentence of the third paragraph of the right column of page 518 which is page 8/9 of the PDF document. The word "historically" is a rewording of the source text's phrase "throughout history". The phrase "appears to be" is a rewording of the source text's word "seem". The phrase "results in a lot of difficulty finding" is a rewording of the source text's phrase "make it very hard to find". The phrase "assist in differentiating" is a rewording of the source text's phrase "help differentiate".--><ref>{{Cite journal |vauthors=Kim JJ, Verdu P, Pakstis AJ, Speed WC, Kidd JR, Kidd KK |date=October 2005 |title=Use of autosomal loci for clustering individuals and populations of East Asian origin |journal=Human Genetics |volume=117 |issue=6 |pages=511–9 |doi=10.1007/s00439-005-1334-8 |pmid=16028061 |s2cid=15585215}}</ref> | |||
| Over 70% of extant genetic diversity among Koreans can be explained by admixture with ancient South Chinese immigrants, who were related to Iron Age Cambodians.<ref>{{Cite journal |last1=Kim |first1=Jungeun |last2=Jeon |first2=Sungwon |last3=Choi |first3=Jae-Pil |last4=Blazyte |first4=Asta |display-authors=3 |date=2020 |title=The Origin and Composition of Korean Ethnicity Analyzed by Ancient and Present-Day Genome Sequences |journal=Genome Biology and Evolution |volume=12 |issue=5 |pages=553–565 |doi=10.1093/gbe/evaa062 |pmid=32219389 |pmc=7250502 }}</ref> | |||
| Jung Jongsun et al. (2010)<!--The article wrote this person's name as "Jongsun Jung." "Jung Jongsun" is this person's name in the Korean name ordering.--> used the following Korean samples for a study: Southeast Korean (sample regions: ], ] and ]), Middle West Korean (sample regions: ], ], ] and ]) and Southwest Korean (sample regions: ], ] and ]).<!--The abbreviations used in the study for the Korean populations (SE Korea, MW Korea & SW Korea) are located in Table 1 which is on page 3/8 of the PDF document.--> Due to political reasons, the study said that it did not use North Korean samples, but the study said that the "''historical migration event of ] from ] Empire (]–AD 668) in Northern Korea imply that Northern lineages remain in South Korea.''"<!--The preceding information is in the first and second sentences of the second paragraph of the left column of page 5/8 of the PDF document.--> The study said that the "''Northern people of the Goguryeo Empire''" are closely related to ], and the study said that this group of people ruled most of Southwest Korea.<!--The preceding information is in the first and second sentences of the first paragraph of the right column of page 2/8 of the PDF document.--> The study said that "''some of the royal families and their subjects in the Goguryeo Empire moved to this region and formed the Baekje Empire in ]–].''" Southwest Koreans are closer to Mongolians in the study's ] than the other two Korean regions in the study are close to Mongolians. Southwest Koreans also display genetic connections with the ] sample of Japanese in ],<!--The preceding information is in the second, third and fourth sentences of the second paragraph of the right column of page 3/8 of the PDF document. The acronym "JPT" refers to the HapMap sample of Japanese in Tokyo, Japan. The last sentence of the second paragraph of the left column of page 3/8 of the PDF document says "JPT (Tokyo)," and Table 1 of the same page says "Japanese HapMap(JPT)."--> and, in the ] tree, the ] for Southwest Korea is close to Japan.<!--The preceding information is in the last sentence of the first paragraph of the right column of page 3/8 of the PDF document. The abbreviation "NJ" used in that sentence was defined as "Neighbor Joining" in the last sentence of the second paragraph of the right column of page 2/8 of the PDF document.--> In the study's Korea-China-Japan genome map, some Southwest Korean samples overlap with samples from Japan.<!--The preceding information is in the first and third sentences of the first paragraph of the right column of page 3/8 of the PDF document.--> The study said that the fairly close relationship, in both the study's ] analysis and ], of the ] Southwest Korean sample and the HapMap sample of Japanese in Tokyo, Japan, has made the evolutionary relationship of Chinese, Japanese and Koreans become clearer.<!--The preceding information is in the last sentence of the first paragraph of the left column of page 5/8 of the PDF document. That sentence's reference to "STRUCTURE results" is explained to be an analysis of genetic structure, "In our analysis of genetics structure by STRUCTURE program," in the first sentence of the third paragraph of the left column of page 3/8 of the PDF document.--> Southeast Koreans display some genetic similarities with people of ], Japan, which indicates that there might have been links between these regions. The study said that it is possible that ]s in the ] sample, one of the sampled Southeast Korean regions, and outliers in the Kobe, Japan, sample both have ] lineage due to Southeast Koreans having connections with Siberian lineages with respect to grave patterns and culture.<!--The preceding information is in the fifth and sixth sentences of the second paragraph of the right column of page 3/8 of the PDF document.--> The overall result for the study's Korea-Japan-China genome map indicates that some signals for Siberia remain in Southeast Korea.<!--The preceding information is in the last sentence of the first paragraph of the left column of page 4/8 of the PDF document. The sentence construction in the source text's sentence which used the word "respectively" meant that the signals for Mongolia remain in SW Korea and signals for Siberia remain in SE Korea.--> In contrast to the ] sample, the ] and ] samples, which are both Southeast Korean samples, displayed average signals for the Korean Peninsula.<!--The preceding information is in the second sentence of the first paragraph of the left column of page 4/8 of the PDF document.--> The study said that Middle West Korea was a ] in the Korean Peninsula with people traveling from North to South, South to North, and people traveling from ], including from the ].<!--The preceding information is in the sixth sentence, which starts with "Model III," and the seventh sentences of the first paragraph of the left column of page 4/8 of the PDF document. The sixth sentence uses the term "SanDung peninsula" for the Shandong Peninsula. The seventh sentence uses the term "Peking" for "Beijing".--> ], which included those in the Shandong Peninsula, travelled across the ], and these Western Chinese lived and traded in both China and Korea.<!--The preceding information is in the fourth sentence, the sentence which starts with "Model III," of the first paragraph of the right column of page 2/8 of the PDF document.--> In the study's genome map, Middle West Koreans are close to the ] sample of Han Chinese in ]<!--The preceding information is in the second-to-last sentence of the first paragraph of the left column of page 4/8 of the PDF document. The acronym "CHB" refers to the HapMap sample of Han Chinese in Beijing, China. The last sentence of the second paragraph of the left column of page 3/8 of the PDF document says "CHB (Peking)," and Table 1 of the same page says "Chinese HapMap(CHB)."--> and, in the ] tree, the ] for Middle West Korea is close to China.<!--The preceding information is in the last sentence of the first paragraph of the right column of page 3/8 of the PDF document. The abbreviation "NJ" used in that sentence was defined as "Neighbor Joining" in the last sentence of the second paragraph of the right column of page 2/8 of the PDF document.--> The overall result for the study's Korea-Japan-China genome map indicates that Middle West Korea displays an average signal for South Korea.<!--The preceding information is in the last sentence of the first paragraph of the left column of page 4/8 of the PDF document.--> Chinese people are located between Korean and Vietnamese people in the study's ].<!--The preceding information is in the second-to-last sentence of the first paragraph of the left column of page 5/8 of the PDF document.--><ref name="JungJongsun2010">{{Cite journal |display-authors=6 |vauthors=Jung J, Kang H, Cho YS, Oh JH, Ryu MH, Chung HW, Seo JS, Lee JE, Oh B, Bhak J, Kim HL |date=July 2010 |title=Gene flow between the Korean peninsula and its neighboring countries |journal=PLOS ONE |volume=5 |issue=7 |pages=e11855 |bibcode=2010PLoSO...511855J |doi=10.1371/journal.pone.0011855 |pmc=2912326 |pmid=20686617 |doi-access=free}}</ref><!--The Coriell Institute citation is being used to cite that the acronym "CHB," which was used in Jung Jongsun et al. (2010) in reference to the Chinese HapMap sample from Beijing, was a sample of Han Chinese.--><ref>{{Cite web |date=2018 |title=Han Chinese in Beijing, China . |url=https://www.coriell.org/1/NHGRI/Collections/HapMap-Collections/Han-Chinese-in-Beijing-China-CHB |access-date=27 February 2018 |website=Coriell Institute for Medical Research}}</ref> | |||
| ==== Paternal lineages ==== | |||
| Kim Young-jin and Jin Han-jun (2013)<!--The study wrote their names as "Young Jin Kim & Han Jun Jin." Kim Young-jin and Jin Han-jun are their names written in the Korean name ordering.--> said that ] had Korean ] samples clustering with neighboring East Asian populations which were geographically nearby them such as the Chinese and Japanese.<!--This information is in the fifth sentence of the third paragraph of the left column of page 6/12 (page 357). Information about the principal component analysis being performed is in the first sentence of that paragraph.--> The study said that Koreans are genetically closely related to ] in comparison to Koreans' genetic relatedness to other East Asians which included the following East and Southeast Asian peoples: ], ], ], ], ], ], ], ], ], ], Southern Han Chinese, Northern Han Chinese, ], ], ], ] and ]. The study said that the close genetic relatedness of Koreans to Japanese has been reported in the following previous studies: Kivisild et al. (2002); Jin et al. (2003); Jin et al. (2009); and Underhill and Kivisild (2007).<!--The preceding information is in the fifth sentence which starts with "On comparison" of the first paragraph of the left column of page 9/12 of the PDF document (page 360). The other East Asian populations which were compared are listed in Table 2 of page 8/12 of the PDF document (page 359).--> The study said that Jung et al. (2010) said that there is a genetic substructure in Koreans, but the study said that it found Korean ] individuals to be highly genetically similar.<!--The preceding information is in the third-to-last sentence which starts with "Genetic sub-structure" of the first paragraph of the right column of page 9/12 of the PDF document (page 360).--> The study said that Jin et al. (2009) found that Koreans from different populations are not different in a significant way which indicates that Koreans are genetically ].<!--The preceding information is in the last sentence of the first paragraph of the right column of page 9/12 of the PDF document (page 360).--> The study said that the affinity of Koreans is predominately Southeast Asian with an estimated admixture of 79% Southeast Asian and 21% Northeast Asian Koreans, but the study said that this does not mean that Koreans are ], because all of the Koreans which were analyzed uniformly displayed a dual pattern of Northeast Asian and Southeast Asian origins.<!--The preceding information is in the second, third and fourth sentences of the second paragraph of the right column of page 9/12 of the PDF document (page 360). On page 10/12 of the PDF document (page 361), Table 3 which lists Koreans' 79% Southeast Asian and 21% Northeast Asian estimated admixture uses the terms "Northeast Asians" and "Southeast Asian" in its title.--> The study said that Koreans and Japanese displayed no observable difference between each other in their proportion of Southeast Asian and Northeast Asian admixture.<!--The preceding information is in the second, third and fourth sentences of the second paragraph of the right column of page 6/12 of the PDF document (page 357). Figure 2 B on page 10/12 of the PDF document (page 361) shows the results referred to in those sentences.--> The study said the 79% Southeast Asian and 21% Northeast Asian admixture estimate for Koreans is consistent with the interpretation of Jin et al. (2009) that Koreans descend from a Northeast Asian population which was subsequently followed by a male-centric migration from the southern region of Asia which changed both the ] composition and ] in the Korean population.<!--The preceding information is in the first and second sentences of the first paragraph of the left column of page 10/12 (page 361).--> | |||
| Studies of ] have so far produced evidence to suggest that the ] have a long history as a distinct, mostly ] ethnic group, with successive waves of people moving to the peninsula and three major Y-chromosome haplogroups.<ref>{{Cite journal |vauthors=Kim SH, Han MS, Kim W, Kim W |date=November 2010 |title=Y chromosome homogeneity in the Korean population |journal=International Journal of Legal Medicine |volume=124 |issue=6 |pages=653–7 |doi=10.1007/s00414-010-0501-1 |pmid=20714743 |s2cid=27125545}}</ref> A majority of Koreans belong to subclades of ] (''ca.'' 79% in total,<ref name="Kim2011" /><ref name="Kwon2015" /> with about 42%<ref name="Kwon2015" /> to 44%<ref name="Kim2011" /> belonging to ], about 31%<ref name="Kim2011" /> to 32%<ref name="Kwon2015" /> belonging to ], and about 2%<ref name="Kim2011" /> to 3%<ref name="Kwon2015" /> belonging to ]), while a significant minority belong to subclades of ] (''ca.'' 12%<ref name="Kim2011" /> to 13%<ref name="Kwon2015" /> in total). Other Y-DNA haplogroups, including ], ], and ], are also found in smaller proportions of present-day Koreans.<ref>{{Cite journal |last1=Zhang |first1=Ai Hua |last2=Lee |first2=Hye Young |last3=Seo |first3=Seung Bum |last4=Lee |first4=Hyo Jung |last5=Jin |first5=Hong Xuan |last6=Cho |first6=So Hee |last7=Lyoo |first7=Sung Hee |last8=Kim |first8=Ki Ha |last9=Lee |first9=Jae Won |last10=Lee |first10=Soong Deok |date=29 May 2012 |title=Y Haplogroup Distribution in Korean and Other Populations |url=https://synapse.koreamed.org/articles/1004691 |journal=Korean Journal of Legal Medicine |volume=36 |issue=1 |pages=34–44 |doi=10.7580/KoreanJLegMed.2012.36.1.34|doi-access=free}}</ref><ref>{{Cite journal |last1=Choi |first1=Sun Seong |last2=Park |first2=Kyung Hwa |last3=Nam |first3=Da Eun |last4=Kang |first4=Tae Hoon |last5=Chung |first5=Ki Wha |date=1 December 2017 |title=Y-chromosome haplogrouping for Asians using Y-SNP target sequencing |journal=Forensic Science International: Genetics Supplement Series |volume=6 |pages=e235–e237 |doi=10.1016/j.fsigss.2017.09.100 |issn=1875-1768|doi-access=free}}</ref><ref>{{Cite journal |last1=Kim |first1=Soon Hee |last2=Han |first2=Myun Soo |last3=Kim |first3=Wook |last4=Kim |first4=Won |date=November 2010 |title=Y chromosome homogeneity in the Korean population |url=https://pubmed.ncbi.nlm.nih.gov/20714743/ |journal=International Journal of Legal Medicine |volume=124 |issue=6 |pages=653–657 |doi=10.1007/s00414-010-0501-1 |issn=1437-1596 |pmid=20714743|s2cid=27125545}}</ref> | |||
| ==== Maternal lineages ==== | |||
| == Genetic history of Mongolic peoples == | |||
| Studies of Korean ] lineages have shown that there is a high frequency of ], followed by ], and then ] and ]. Haplogroups with lower frequency include ], ], ], ], ], ], ], ], ], ], ], and ].<ref>{{Cite journal |last1=Jin |first1=Han-Jun |last2=Tyler-Smith |first2=Chris |last3=Kim |first3=Wook |date=16 January 2009 |title=The Peopling of Korea Revealed by Analyses of Mitochondrial DNA and Y-Chromosomal Markers |journal=PLOS ONE |volume=4 |issue=1 |pages=e4210 |doi=10.1371/journal.pone.0004210 |issn=1932-6203 |pmc=2615218 |pmid=19148289 |bibcode=2009PLoSO...4.4210J |doi-access=free}}</ref><ref>{{Cite journal |last1=Jeon |first1=Sungwon |last2=Bhak |first2=Youngjune |last3=Choi |first3=Yeonsong |last4=Jeon |first4=Yeonsu |last5=Kim |first5=Seunghoon |last6=Jang |first6=Jaeyoung |last7=Jang |first7=Jinho |last8=Blazyte |first8=Asta |last9=Kim |first9=Changjae |last10=Kim |first10=Yeonkyung |last11=Shim |first11=Jungae |last12=Kim |first12=Nayeong |last13=Kim |first13=Yeo Jin |last14=Park |first14=Seung Gu |last15=Kim |first15=Jungeun |date=27 May 2020 |title=Korean Genome Project: 1094 Korean personal genomes with clinical information |journal=Science Advances |volume=6 |issue=22 |pages=eaaz7835 |doi=10.1126/sciadv.aaz7835 |issn=2375-2548 |pmc=7385432 |pmid=32766443|bibcode=2020SciA....6.7835J}}</ref><ref>{{Cite journal |last1=Hong |first1=Seung Beom |last2=Kim |first2=Ki Cheol |last3=Kim |first3=Wook |date=1 October 2014 |title=Mitochondrial DNA haplogroups and homogeneity in the Korean population |url=https://doi.org/10.1007/s13258-014-0194-9 |journal=Genes & Genomics |language=en |volume=36 |issue=5 |pages=583–590 |doi=10.1007/s13258-014-0194-9 |s2cid=256067815 |issn=2092-9293}}</ref> | |||
| {{See also|Mongolic peoples}} | |||
| === Mongolic peoples === | |||
| The ] are an ethnic group in northern China, Mongolia, parts of Siberia and Western Asia. They are believed to be the descendants of the ] and the ]. The former term includes the Mongols proper (also known as the ]), ], the ] and the Southern Mongols. The latter comprises the ], ], ], ], ], ], ], ], ], ] and ]. The ] are descendants of the para-Mongolic ].<ref>{{Cite web |title=DNA Match Solves Ancient Mystery |url=http://china.org.cn/english/2001/Aug/16896.htm |access-date=25 December 2018 |website=china.org.cn}}</ref> Mongolians are also related to the Manchurians. | |||
| {{Main|Mongolic peoples}} | |||
| The ethnogenesis of Mongolic peoples is largely linked with the expansion of ]s. They subsequently came into contact with other groups, notably ] to their South and ] to their far West. The Mongolians pastoralist lifestyle, may in part be derived from the Western Steppe Herders, but without much geneflow between these two groups, suggesting cultural transmission.<ref>{{Cite web |title=Population dynamics and the rise of empires in Inner Asia: Genome-wide analysis spanning 6,000 years in the eastern Eurasian Steppe gives insights to the formation of Mongolia's empires |url=https://www.sciencedaily.com/releases/2020/11/201105183836.htm |access-date=4 June 2023 |website=ScienceDaily |language=en}}</ref><ref>{{Cite journal |last1=Yang |first1=Xiaomin |last2=Sarengaowa |last3=He |first3=Guanglin |last4=Guo |first4=Jianxin |last5=Zhu |first5=Kongyang |last6=Ma |first6=Hao |last7=Zhao |first7=Jing |last8=Yang |first8=Meiqing |last9=Chen |first9=Jing |last10=Zhang |first10=Xianpeng |last11=Tao |first11=Le |last12=Liu |first12=Yilan |last13=Zhang |first13=Xiu-Fang |last14=Wang |first14=Chuan-Chao |date=2021 |title=Genomic Insights Into the Genetic Structure and Natural Selection of Mongolians |journal=Frontiers in Genetics |volume=12 |page=735786 |doi=10.3389/fgene.2021.735786 |issn=1664-8021 |pmc=8693022 |pmid=34956310 |doi-access=free}}</ref> The ] are believed to be the descendants of the ] and the ]. The former term includes the Mongols proper (also known as the ]), ], the ] and the Southern Mongols. The latter comprises the ], ], ], ], ], ], ], ], ], ] and ]. The ] are descendants of the para-Mongolic ].<ref>{{Cite web |title=DNA Match Solves Ancient Mystery |url=http://china.org.cn/english/2001/Aug/16896.htm |access-date=25 December 2018 |website=china.org.cn}}</ref> | |||
| === Paternal lineages === | |||
| The majority of Mongols in ] and ] belong to Y-DNA ]. Haplogroup C-M217 among the Mongols is characterized by a diversity that dates back to the very origin of haplogroup C-M217 and very shallow diversity in each of the frequently observed subclades: C-M504, ], C-M407, and C-F1756. Of these four subclades, C-M407 is phylogenetically extremely divergent from the others, and is more closely related to subclades of C-M217 that are found among present-day Chinese, Koreans, Japanese, and other East and Southeast Asians; however, among Mongols, C-M407 is found most frequently in northern tribes<ref name="Malyarchuk2016">{{Cite journal |vauthors=Malyarchuk BA, Derenko M, Denisova G, Woźniak M, Rogalla U, Dambueva I, Grzybowski T |date=June 2016 |title=Y chromosome haplotype diversity in Mongolic-speaking populations and gene conversion at the duplicated STR DYS385a,b in haplogroup C3-M407 |journal=Journal of Human Genetics |volume=61 |issue=6 |pages=491–496 |doi=10.1038/jhg.2016.14 |pmid=26911356 |doi-access=free |s2cid=13217444}}</ref> and Buryats<ref name="pmid25711029">{{Cite journal |vauthors=Har'kov VN, Hamina KV, Medvedeva OF, Simonova KV, Eremina ER, Stepanov VA |date=February 2014 |title= |journal=Genetika |language=ru |volume=50 |issue=2 |pages=203–213 |doi=10.1134/S1022795413110082 |pmid=25711029 |s2cid=15595963}}</ref><ref name="Malyarchuk2010">{{Cite journal |vauthors=Malyarchuk B, Derenko M, Denisova G, Wozniak M, Grzybowski T, Dambueva I, Zakharov I |date=November 2010 |title=Phylogeography of the Y-chromosome haplogroup C in northern Eurasia |journal=Annals of Human Genetics |volume=74 |issue=6 |pages=539–546 |doi=10.1111/j.1469-1809.2010.00601.x |pmid=20726964 |s2cid=40763875|doi-access=free }}</ref>) and toward the west (among Dorbet Kalmyks<ref>{{Cite journal |vauthors=Malyarchuk B, Derenko M, Denisova G, Khoyt S, Woźniak M, Grzybowski T, Zakharov I |date=December 2013 |title=Y-chromosome diversity in the Kalmyks at the ethnical and tribal levels |journal=Journal of Human Genetics |volume=58 |issue=12 |pages=804–811 |doi=10.1038/jhg.2013.108 |pmid=24132124 |doi-access=free}}</ref><ref name="Balinova2019">{{Cite journal |display-authors=6 |vauthors=Balinova N, Post H, Kushniarevich A, Flores R, Karmin M, Sahakyan H, Reidla M, Metspalu E, Litvinov S, Dzhaubermezov M, Akhmetova V, Khusainova R, Endicott P, Khusnutdinova E, Orlova K, Bakaeva E, Khomyakova I, Spitsina N, Zinchenko R, Villems R, Rootsi S |date=September 2019 |title=Y-chromosomal analysis of clan structure of Kalmyks, the only European Mongol people, and their relationship to Oirat-Mongols of Inner Asia |journal=European Journal of Human Genetics |volume=27 |issue=9 |pages=1466–1474 |doi=10.1038/s41431-019-0399-0 |pmc=6777519 |pmid=30976109}}</ref>). | |||
| ==== Paternal lineages ==== | |||
| ] and ] are found at medium rates among present-day Mongols. The subclades of Haplogroup O-M175 that have been observed among Mongols tend to be similar to those found among Han Chinese; in fact, a plurality of ] have been found to belong to ], just like the Han Chinese majority in China.<ref name = "He2022">Guang‐Lin He, Meng‐Ge Wang, Xing Zou, Hui‐Yuan Yeh, Chang‐Hui Liu, Chao Liu, Gang Chen, and Chuan‐Chao Wang. "Extensive ethnolinguistic diversity at the crossroads of North China and South Siberia reflects multiple sources of genetic diversity." ''J Syst Evol'', 2023, 61(1): 230-250. https://doi.org/10.1111/jse.12827</ref> The subclades of Haplogroup N-M231 that have been observed among Mongols in Mongolia and Russia tend to carry either the Siberian N-B478 clade, or N-B197, a Far Eastern clade.<ref name="Ilumae2016">{{cite journal | vauthors = Ilumäe AM, Reidla M, Chukhryaeva M, Järve M, Post H, Karmin M, Saag L, Agdzhoyan A, Kushniarevich A, Litvinov S, Ekomasova N, Tambets K, Metspalu E, Khusainova R, Yunusbayev B, Khusnutdinova EK, Osipova LP, Fedorova S, Utevska O, Koshel S, Balanovska E, Behar DM, Balanovsky O, Kivisild T, Underhill PA, Villems R, Rootsi S | display-authors = 6 | title = Human Y Chromosome Haplogroup N: A Non-trivial Time-Resolved Phylogeography that Cuts across Language Families | journal = American Journal of Human Genetics | volume = 99 | issue = 1 | pages = 163–173 | date = July 2016 | pmid = 27392075 | pmc = 5005449 | doi = 10.1016/j.ajhg.2016.05.025 }}</ref> Among Mongols in China, typically Chinese subclades of haplogroup N, have been found more often than the aforementioned typically North Asian subclades of haplogroup N.<ref name = "He2022" /> In addition, some members of a wide variety of other Y-DNA haplogroups have been found among present-day Mongols, including ], and a variety of West Eurasian haplogroups.<ref>{{Cite web |title=Mongol Genetics – DNA of Mongolia's Khalkha Mongolians and others |url=http://www.khazaria.com/genetics/mongols.html |access-date=25 December 2018 |website=khazaria.com}}</ref> | |||
| The majority of Mongols in ] and ] belong to subclades of ],<ref>{{Cite journal |vauthors=Malyarchuk BA, Derenko M, Denisova G, Woźniak M, Rogalla U, Dambueva I, Grzybowski T |date=June 2016 |title=Y chromosome haplotype diversity in Mongolic-speaking populations and gene conversion at the duplicated STR DYS385a,b in haplogroup C3-M407 |journal=Journal of Human Genetics |volume=61 |issue=6 |pages=491–496 |doi=10.1038/jhg.2016.14 |pmid=26911356 |doi-access=free |s2cid=13217444}}</ref> followed by lower frequency of ] and ].<ref>Guang‐Lin He, Meng‐Ge Wang, Xing Zou, Hui‐Yuan Yeh, Chang‐Hui Liu, Chao Liu, Gang Chen, and Chuan‐Chao Wang. "Extensive ethnolinguistic diversity at the crossroads of North China and South Siberia reflects multiple sources of genetic diversity." ''J Syst Evol'', 2023, 61(1): 230-250. https://doi.org/10.1111/jse.12827</ref> A minority belongs to ], and a variety of West Eurasian haplogroups.<ref>{{Cite web |title=Mongol Genetics – DNA of Mongolia's Khalkha Mongolians and others |url=http://www.khazaria.com/genetics/mongols.html |access-date=25 December 2018 |website=khazaria.com}}</ref> | |||
| === Maternal lineages === | ==== Maternal lineages ==== | ||
| The maternal haplogroups are diverse but similar to other northern Asian populations, including ], ], ], and ], which are shared among indigenous American and Asian populations.<ref>{{Cite journal |vauthors=Kolman CJ, Sambuughin N, Bermingham E |date=April 1996 |title=Mitochondrial DNA analysis of Mongolian populations and implications for the origin of New World founders |journal=Genetics |volume=142 |issue=4 |pages=1321–34 |doi=10.1093/genetics/142.4.1321 |pmc=1207128 |pmid=8846908}}</ref> West Eurasian mtDNA haplogroups makes up a some minority percentages. ], ], ], ], ] are all found in Mongolic people.<ref>{{Cite journal |display-authors=6 |vauthors=Cheng B, Tang W, He L, Dong Y, Lu J, Lei Y, Yu H, Zhang J, Xiao C |date=October 2008 |title=Genetic imprint of the Mongol: signal from phylogeographic analysis of mitochondrial DNA |journal=Journal of Human Genetics |volume=53 |issue=10 |pages=905–913 |doi=10.1007/s10038-008-0325-8 |pmid=18769869 |quote=European-prevalent haplogroups (HV, U, K, I, J) are 14.3% in Xinjiang Mongolian, 10% in Mongolia, 8.4% in central Inner Mongolian samples, and only 2% in eastern Xin Barage Zuoqi County samples, showing decreasing frequencies from west to east |doi-access=free}}</ref> | |||
| === Han Chinese === | |||
| The maternal haplogroups are diverse but similar to other northern Asian populations, including ], ], ], and ], which are shared among indigenous American and Asian populations.<ref>{{Cite journal |vauthors=Kolman CJ, Sambuughin N, Bermingham E |date=April 1996 |title=Mitochondrial DNA analysis of Mongolian populations and implications for the origin of New World founders |journal=Genetics |volume=142 |issue=4 |pages=1321–34 |doi=10.1093/genetics/142.4.1321 |pmc=1207128 |pmid=8846908}}</ref> | |||
| {{See also|Han Chinese}}]The origins of the Han Chinese are primarily from Neolithic Yellow River farmers, which formed from ]s (ANEA) and Neolithic groups near the ], which formed ]s (ASEA).<ref name=":4">{{cite journal |author1=Wang, Yuchen |author2=Lu Dongsheng |author3=Chung Yeun-Jun |author4=Xu Shuhua |year=2018 |title=Genetic structure, divergence and admixture of Han Chinese, Japanese and Korean populations |journal=Hereditas |volume=155 |page=19 |doi=10.1186/s41065-018-0057-5 |pmc=5889524 |pmid=29636655 |doi-access=free}}</ref><ref name=":7" /><ref>{{cite journal |last1=Cao |first1=Yanan |last2=Li |first2=Lin |last3=Xu |first3=Min |display-authors=etal |date=2020 |title=The ChinaMAP analytics of deep whole genome sequences in 10,588 individuals |journal=Cell Research |volume=30 |issue=9 |pages=717–731 |doi=10.1038/s41422-020-0322-9 |pmc=7609296 |pmid=32355288 |doi-access=free}}</ref><ref name=":8">{{Cite journal |last1=Pan |first1=Ziqing |last2=Xu |first2=Shuhua |date=2019 |title=Population genomics of East Asian ethnic groups |journal=] |location=Berlin |publisher=] |publication-date=2020 |volume=157 |issue=49 |page=49 |doi=10.1186/s41065-020-00162-w |pmc=7724877 |pmid=33292737 |doi-access=free}}</ref><ref>{{Cite journal |last1=Shi |first1=Cheng-Min |last2=Liu |first2=Qi |last3=Zhao |first3=Shilei |last4=Chen |first4=Hua |date=21 March 2019 |title=Ancestry informative SNP panels for discriminating the major East Asian populations: Han Chinese, Japanese and Korean |url=https://www.researchgate.net/publication/332249278 |url-status=live |journal=] |location=Cambridge |publisher=] |publication-date=21 March 2019 |volume=29 |issue=2 |pages=348–354 |doi=10.1111/ahg.12320 |pmid=31025319 |archive-url=https://web.archive.org/web/20211105170923/https://www.researchgate.net/publication/332249278_Ancestry_informative_SNP_panels_for_discriminating_the_major_East_Asian_populations_Han_Chinese_Japanese_and_Korean |archive-date=5 November 2021 |access-date=14 September 2021 |doi-access=free}}</ref> Today's modern Han Chinese can be categorized out of convenience into two subgroups, Northern and Southern Han Chinese, despite there being no clear genetic divide between the north and south because the Han Chinese are a clinal population.<ref name=zhaoyong-bin/> The Northern Han Chinese carry mostly ANEA ancestry with some degree of ASEA admixture, whereas the Southern Han Chinese carry significantly higher levels of ASEA ancestry than Northern Han Chinese, although ANEA ancestry is still significant.{{Citation needed|date=January 2025}}<ref name=":7">{{Cite journal |last1=Yang |first1=Melinda A. |last2=Fan |first2=Xuechun |last3=Sun |first3=Bo |last4=Chen |first4=Chungyu |last5=Lang |first5=Jianfeng |last6=Ko |first6=Ying-Chin |last7=Tsang |first7=Cheng-hwa |last8=Chiu |first8=Hunglin |last9=Wang |first9=Tianyi |last10=Bao |first10=Qingchuan |last11=Wu |first11=Xiaohong |last12=Hajdinjak |first12=Mateja |last13=Ko |first13=Albert Min-Shan |last14=Ding |first14=Manyu |last15=Cao |first15=Peng |date=17 July 2020 |title=Ancient DNA indicates human population shifts and admixture in northern and southern China |url=https://www.science.org/doi/10.1126/science.aba0909 |journal=Science |language=en |volume=369 |issue=6501 |pages=282–288 |doi=10.1126/science.aba0909 |pmid=32409524 |bibcode=2020Sci...369..282Y |s2cid=218649510 |issn=0036-8075}}</ref><ref name="sciencedirect.com" /><ref>{{Cite journal |last=Ebrey |first=Patricia Buckley |date=2023 |title=Rethinking Han Chinese Identity |url=https://www.jstor.org/stable/48726991 |journal=China Review |volume=23 |issue=2 |pages=57–86 |jstor=48726991 |issn=1680-2012}}</ref> In large, the Han Chinese cluster retains a level of singularity with its admixture of ANEA and ASEA ancestries which is unique to the group.<ref name=":8" /> When comparing with other East Asian populations, the Northern Han Chinese cluster is related to the "]/] cluster" in terms of a correlative genetic relationship (mostly due to the overlap of ANEA), but is also quite distinguishable from them genetically,<ref name=":4" /><ref name=":9">{{Citation |last=Huang |first=Xiufeng |title=Genomic Insights into the Demographic History of Southern Chinese |date=8 November 2020 |url=https://www.biorxiv.org/content/10.1101/2020.11.08.373225v1 |access-date= |language=en |doi=10.1101/2020.11.08.373225 |last2=Xia |first2=Zi-Yang |last3=Bin |first3=Xiaoyun |last4=He |first4=Guanglin |last5=Guo |first5=Jianxin |last6=Lin |first6=Chaowen |last7=Yin |first7=Lianfei |last8=Zhao |first8=Jing |last9=Ma |first9=Zhuofei}}</ref> due to the presence of ASEA ancestry<ref>{{Cite journal |last1=Feng |first1=Qidi |last2=Lu |first2=Yan |last3=Ni |first3=X. |last4=Yuan |first4=K. |last5=Yang |first5=Ya-jun |last6=Yang |first6=Xiong |last7=Liu |first7=Chang |last8=Lou |first8=H. |last9=Ning |first9=Zhilin |last10=Wang |first10=Yuchen |last11=Lu |first11=Dongsheng |last12=Zhang |first12=Chao |last13=Zhou |first13=Ying |last14=Shi |first14=Meng |last15=Tian |first15=L. |date=2017 |title=Genetic History of Xinjiang's Uyghurs Suggests Bronze Age Multiple-Way Contacts in Eurasia |journal=Molecular Biology and Evolution |language=en |volume=34 |issue=10 |pages=2572–2582 |doi=10.1093/molbev/msx177 |pmid=28595347 |s2cid=28730957 |doi-access=free}}</ref> and the absence of Jōmon ancestry. The Southern Han Chinese also share more alleles with ] and other Kra–Dai peoples according to ] than Northern Han Chinese.<ref name=":9" /> | |||
| The genetic makeup of the modern Han Chinese is not purely uniform in terms of physical appearance and biological structure due to the vast geographical expanse of China and the ] that have occurred throughout it over the last few millennia. This has also engendered the emergence and evolution of the diverse multiplicity of assorted ] found throughout the various regions of modern China today. Comparisons between the Y chromosome ]s (SNPs) and ] (mtDNA) of modern Northern Han Chinese and 3000 year old Hengbei ancient samples from ] show that they are extremely similar to each other. These findings demonstrate that the core fundamental structural basis that shaped the genetic makeup of the present-day Northern Han Chinese was already formed more than three thousand years ago.<ref name=zhaoyong-bin>{{cite journal |last1=Zhao |first1=Yong-Bin |last2=Zhang |first2=Ye |last3=Zhang |first3=Quan-Chao |last4=Li |first4=Hong-Jie |last5=Cui |first5=Ying-Qiu |last6=Xu |first6=Zhi |last7=Jin |first7=Li |last8=Zhou |first8=Hui |last9=Zhu |first9=Hong |editor1-last=Hofreiter |editor1-first=Michael |title=Ancient DNA Reveals That the Genetic Structure of the Northern Han Chinese Was Shaped Prior to 3,000 Years Ago |journal=PLOS ONE |date=4 May 2015 |volume=10 |issue=5 |pages=e0125676 |doi=10.1371/journal.pone.0125676 |pmid=25938511 |pmc=4418768 |bibcode=2015PLoSO..1025676Z |doi-access=free}}</ref> | |||
| Derenko ''et al.'' (2007) tested a sample of 47 Mongolians from Ulaanbaatar and found that they were 17.0% ], 14.9% ], 12.8% ], 10.6% ], 10.6% ], 8.5% ], 6.4% haplogroup U4 and 2.1% haplogroup K), 6.4% ], 4.3% haplogroup M8a2, 2.1% haplogroup M7, 2.1% haplogroup M9a, 2.1% haplogroup M10, 2.1% ], 2.1% haplogroup M*, 2.1% haplogroup N9a, 2.1% haplogroup N1.<ref name="Derenko2007"/> | |||
| Studies of DNA remnants from the Central Plains area of China 3000 years ago show close affinity between that population and those of Northern Han today in both the Y-DNA and mtDNA. Both northern and southern Han show similar Y-DNA genetic structure.<ref>{{cite journal |last1=Zhao |first1=Yong-Bin |last2=Zhang |first2=Ye |last3=Zhang |first3=Quan-Chao |last4=Li |first4=Hong-Jie |last5=Cui |first5=Ying-Qiu |last6=Xu |first6=Zhi |last7=Jin |first7=Li |last8=Zhou |first8=Hui |last9=Zhu |first9=Hong |title=Ancient DNA Reveals That the Genetic Structure of the Northern Han Chinese Was Shaped Prior to 3,000 Years Ago |journal=PLOS ONE |date=4 May 2015 |volume=10 |issue=5 |pages=e0125676 |doi=10.1371/journal.pone.0125676 |pmid=25938511 |pmc=4418768 |bibcode=2015PLoSO..1025676Z |doi-access=free}}</ref> | |||
| Jin ''et al.'' (2009) tested another sample of 47 Mongolians from Ulaanbaatar and found that they were 10/47 = 21.3% ], 17.0% ] 14.9% ] 12.8% ] 8.5% ], 4.3% ], 4.3% haplogroup M7, 4.3% haplogroup N9a, 2.1% haplogroup M*, 2.1% haplogroup M8a, 2.1% haplogroup U5a, 2.1% haplogroup Y1, 2.1% haplogroup pre-Z, and 2.1% ].<ref name="Jin2009">{{Cite journal |vauthors=Jin HJ, Tyler-Smith C, Kim W |year=2009 |title=The peopling of Korea revealed by analyses of mitochondrial DNA and Y-chromosomal markers |journal=PLOS ONE |volume=4 |issue=1 |pages=e4210 |bibcode=2009PLoSO...4.4210J |doi=10.1371/journal.pone.0004210 |pmc=2615218 |pmid=19148289 |doi-access=free}}</ref> | |||
| Northern Han Chinese populations also have some West Eurasian admixture,<ref>{{Cite journal |last1=Chiang |first1=Charleston W.K. |last2=Mangul |first2=Serghei |last3=Robles |first3=Christopher |last4=Sankararaman |first4=Sriram |date=2018 |title=A Comprehensive Map of Genetic Variation in the World's Largest Ethnic Group —Han Chinese |journal=Molecular Biology and Evolution |volume=35 |issue=11 |pages=2736–2750 |doi=10.1093/molbev/msy170 |pmid=30169787 |pmc=6693441 }}</ref> especially Han Chinese populations in ] (~2%-4.6%)<ref>{{Cite journal |last1=He |first1=Guang-Lin |last2=Wang |first2=Meng-Ge |last3=Li |first3=Ying-Xiang |last4=Zou |first4=Xing |display-authors=3 |date=2022 |title=Fine-scale north-to-south genetic admixture profile in Shaanxi Han Chinese revealed by genome-wide demographic history reconstruction |url=https://onlinelibrary.wiley.com/doi/abs/10.1111/jse.12715 |journal=Journal of Systematics and Evolution |volume=60 |issue=4 |pages=955–972 |doi=10.1111/jse.12715 |via=Wiley Online Library}}</ref> and ] (~2%).<ref>{{Cite journal |last1=Zhou |first1=Jingbin |last2=Zhang |first2=Xianpeng |last3=Li |first3=Xin |last4=Sui |first4=Jie |display-authors=3 |date=2022 |title=Genetic structure and demographic history of Northern Han people in Liaoning Province inferred from genome-wide array data |journal=Frontiers in Ecology and Evolution |volume=10 |doi=10.3389/fevo.2022.1014024 |doi-access=free }}</ref> During the Zhou dynasty, or earlier, peoples with ] haplogroup Q-M120 also contributed to the ethnogenesis of Han Chinese people. This haplogroup is implied to be widespread in the Eurasian steppe and north Asia since it is found among ] in ] and Bronze Age natives of ]. But it is currently near-absent in these regions except for East Asia. In modern China, haplogroup Q-M120 can be found in the northern and eastern regions.<ref>{{Cite journal |last1=Sun |first1=Na |last2=Ma |first2=Peng-Cheng |last3=Yan |first3=Shi |last4=Wen |first4=Shao-Qing |display-authors=3 |date=2019 |title=Phylogeography of Y-chromosome haplogroup Q1a1a-M120, a paternal lineage connecting populations in Siberia and East Asia |url=https://www.tandfonline.com/doi/full/10.1080/03014460.2019.1632930 |journal=Annals of Human Biology |volume=46 |issue=3 |pages=261–266 |doi=10.1080/03014460.2019.1632930 |pmid=31208219 |via=Taylor & Francis Online}}</ref> | |||
| Bai ''et al.'' (2018) tested a sample of 35 ] from Ulaanbaatar, Mongolia and found that they were 28.6% ], 17.1% ], 11.4% ], 11.4% ], 5.7% ], 5.7% ], 5.7% haplogroup N9/Y, 5.7% haplogroup N/A, 2.9% haplogroup M7, 2.9% haplogroup M9, and 2.9% ].<ref name="Bai2018">{{Cite journal |display-authors=6 |vauthors=Bai H, Guo X, Narisu N, Lan T, Wu Q, Xing Y, Zhang Y, Bond SR, Pei Z, Zhang Y, Zhang D, Jirimutu J, Zhang D, Yang X, Morigenbatu M, Zhang L, Ding B, Guan B, Cao J, Lu H, Liu Y, Li W, Dang N, Jiang M, Wang S, Xu H, Wang D, Liu C, Luo X, Gao Y, Li X, Wu Z, Yang L, Meng F, Ning X, Hashenqimuge H, Wu K, Wang B, Suyalatu S, Liu Y, Ye C, Wu H, Leppälä K, Li L, Fang L, Chen Y, Xu W, Li T, Liu X, Xu X, Gignoux CR, Yang H, Brody LC, Wang J, Kristiansen K, Burenbatu B, Zhou H, Yin Y |date=December 2018 |title=Whole-genome sequencing of 175 Mongolians uncovers population-specific genetic architecture and gene flow throughout North and East Asia |journal=Nature Genetics |volume=50 |issue=12 |pages=1696–1704 |doi=10.1038/s41588-018-0250-5 |pmid=30397334 |s2cid=53222895}}</ref> | |||
| <ref>{{cite journal |doi=10.1086/430051 |title=The Dual Origin of the Malagasy in Island Southeast Asia and East Africa: Evidence from Maternal and Paternal Lineages |year=2005 |last1=Hurles |first1=M |last2=Sykes |first2=B |last3=Jobling |first3=M |last4=Forster |first4=P |journal=The American Journal of Human Genetics |volume=76 |issue=5 |pages=894–901 |pmid=15793703 |pmc=1199379}}</ref> Other Y-DNA haplogroups that have been found with notable frequency in samples of Han Chinese include ] (15/165 = 9.1%, 217/2091 = 10.38%,<ref name="He2023" /> 47/361 = 13.0%), ] (10/168 = 6.0%, 27/361 = 7.5%, 176/2091 = 8.42%,<ref name="He2023" /> 187/1730 = 10.8%, 20/166 = 12.0%), ] (6/166 = 3.6%, 94/2091 = 4.50%,<ref name="He2023" /> 18/361 = 5.0%, 117/1729 = 6.8%, 17/165 = 10.3%), ] (78/2091 = 3.73%,<ref name="He2023" /> 54/1147 = 4.7%,<ref name="Lu2009" /> 8/168 = 4.8%, 23/361 = 6.4%, 12/166 = 7.2%), and ] (2/168 = 1.2%, 49/1729 = 2.8%, 61/2091 = 2.92%,<ref name="He2023" /> 12/361 = 3.3%, 48/1147 = 4.2%<ref name="Lu2009" />). | |||
| However, the mtDNA of Han Chinese increases in diversity as one looks from northern to southern China, which suggests that the influx of male Han Chinese migrants intermarried with the local female non-Han aborigines after arriving in what is now modern-day Guangdong, Fujian, and other regions of southern China.<ref name="table" /><ref>{{cite journal |doi=10.1038/sj.ejhg.5201998 |title=A spatial analysis of genetic structure of human populations in China reveals distinct difference between maternal and paternal lineages |year=2008 |last1=Xue |first1=Fuzhong |last2=Wang |first2=Yi |last3=Xu |first3=Shuhua |last4=Zhang |first4=Feng |last5=Wen |first5=Bo |last6=Wu |first6=Xuesen |last7=Lu |first7=Ming |last8=Deka |first8=Ranjan |last9=Qian |first9=Ji |last10=Jin |first10=L |display-authors=9|journal=European Journal of Human Genetics |volume=16 |issue=6 |pages=705–17 |pmid=18212820|doi-access=free}}</ref> Despite this, tests comparing the genetic profiles of northern Han, southern Han, and non-Han southern natives determined that haplogroups O1b-M110, O2a1-M88 and O3d-M7, which are prevalent in non-Han southern natives, were only observed in some southern Han Chinese (4% on average), but not in the northern Han genetic profile. Therefore, this proves<!--suggests???--> that the male contribution of the southern non-Han natives in the southern Han genetic profile is limited, assuming that the frequency distribution of Y lineages in southern non-Han natives represents that prior to the ].<ref name="table" /><ref name="gene" /> | |||
| West Eurasian mtDNA haplogroups ], ], ], ], ], represents 14% in western Xingjang Mongolian, 10% in Mongolia, 8.4% in central Inner Mongolian samples, 2% in eastern Xin Barage Zuoqi County samples.<ref>{{Cite journal |display-authors=6 |vauthors=Cheng B, Tang W, He L, Dong Y, Lu J, Lei Y, Yu H, Zhang J, Xiao C |date=October 2008 |title=Genetic imprint of the Mongol: signal from phylogeographic analysis of mitochondrial DNA |journal=Journal of Human Genetics |volume=53 |issue=10 |pages=905–913 |doi=10.1007/s10038-008-0325-8 |pmid=18769869 |quote=European-prevalent haplogroups (HV, U, K, I, J) are 14.3% in Xinjiang Mongolian, 10% in Mongolia, 8.4% in central Inner Mongolian samples, and only 2% in eastern Xin Barage Zuoqi County samples, showing decreasing frequencies from west to east |doi-access=free}}</ref> West Eurasian mtDNA haplogroups ], ] represents 14.3% in Khalkha. | |||
| A recent, and to date the most extensive, genome-wide association study of the Han population, shows that geographic-genetic stratification from north to south has occurred and centrally placed populations act as the conduit for outlying ones.<ref>{{cite journal |doi=10.1016/j.ajhg.2009.10.016 |title=Genetic Structure of the Han Chinese Population Revealed by Genome-wide SNP Variation |year=2009 |last1=Chen |first1=Jieming |last2=Zheng |first2=Houfeng |last3=Bei |first3=Jin-Xin |last4=Sun |first4=Liangdan |last5=Jia |first5=Wei-hua |last6=Li |first6=Tao |last7=Zhang |first7=Furen |last8=Seielstad |first8=Mark |last9=Zeng |first9=Yi-Xin |last10=Zhang |first10=X |last11=Liu |first11=J |display-authors=9 |journal=The American Journal of Human Genetics |volume=85 |issue=6 |pages=775–85 |pmid=19944401 |pmc=2790583}}</ref> Ultimately, with the exception in some ] branches of the Han Chinese, such as ] and ],<ref>{{cite journal| doi=10.1016/0035-9203(71)90185-4 | volume=65 | title=Cooley's anaemia among the tanka of South China | year=1971 | journal=Transactions of the Royal Society of Tropical Medicine and Hygiene | pages=59–62 | author=McFadzean A.J.S., Todd D. | pmid=5092429 | issue=1}}</ref> there is a "coherent genetic structure"<!-- "coherent genetic structure" doesn't seem to have a standard, formal definition--> found in the entirety of the modern Han Chinese populace.<ref>{{Cite journal |title=Pinghua population as an exception of Han Chinese's coherent genetic structure |doi=10.1007/s10038-008-0250-x |pmid=18270655 |journal=Journal of Human Genetics|volume=53|issue=4|pages=303–13 |year=2008 |last1=Gan |first1=Rui-Jing |last2=Pan |first2=Shang-Ling |last3=Mustavich |first3=Laura F. |last4=Qin |first4=Zhen-Dong |last5=Cai |first5=Xiao-Yun |last6=Qian |first6=Ji |last7=Liu |first7=Cheng-Wu |last8=Peng |first8=Jun-Hua |last9=Li |first9=Shi-Lin |last10=Xu |first10=Jie-Shun |last11=Jin |first11=Li |last12=Li |first12=Hui |doi-access=free}}</ref> Although admixture proportions can vary according to geographic region, the average genetic distance between various Han Chinese populations is much lower than between European populations, for example.<ref>{{Cite journal |last1=Yang |first1=Xiaomin |last2=Wang |first2=Xiao-Xun |last3=He |first3=Guanglin |last4=Guo |first4=Jianxin |display-authors=3 |date=2020 |title=Genomic insight into the population history of Central Han Chinese |url=https://www.researchgate.net/publication/346074810 |journal=Annals of Human Biology |via=ResearchGate}}</ref> | |||
| == Genetic history of Han Chinese == | |||
| {{See also|Han Chinese}} | |||
| ==== Autosomal DNA ==== | |||
| ] | |||
| A 2018 study calculated pairwise F<sub>ST</sub> (a measure of genetic difference) based on genome-wide SNPs, among the Han Chinese (Northern Han from ] and Southern Han from ], ] and ] provinces), Japanese and Korean populations sampled. It found that the smallest F<sub>ST</sub> value was between Northern Han Chinese (Beijing) (CHB) and Southern Han (Hunan, Fujian, etc.) Chinese (CHS) (F<sub>ST</sub> = 0.0014), while CHB and Korean (KOR) (F<sub>ST</sub> = 0.0026) and between KOR and Japanese (JPT) (F<sub>ST</sub> = 0.0033). Generally, pairwise F<sub>ST</sub> between Han Chinese, Japanese and Korean (0.0026~ 0.0090) are greater than that within Han Chinese (0.0014). These results suggested Han Chinese, Japanese and Korean are different in terms of genetic make-up, and the differences among the three groups are much larger than that between northern and southern Han Chinese.<ref>{{cite journal |vauthors=Wang Y, Lu D, Chung YJ, Xu S |title=Genetic structure, divergence and admixture of Han Chinese, Japanese and Korean populations |journal=Hereditas |volume=155 |issue=1 |pages=19 |date=6 April 2018 |pmid=29636655 |pmc=5889524 |doi=10.1186/s41065-018-0057-5 |doi-access=free}} *{{cite web | vauthors = Xu S |date=10 April 2018 |title=Common ancestor of Han Chinese, Japanese and Koreans dated to 3000 – 3600 years ago |url=http://blogs.biomedcentral.com/on-biology/2018/04/10/common-ancestor-of-han-chinese-japanese-and-koreans-dated-to-3000-3600-years-ago/ |website=Biomed Central}}</ref> Nonetheless, there is also genetic diversity among the Southern Han Chinese. The genetic composition of the Han population in Fujian might not accurately represent that of the Han population in Guangdong. | |||
| ] | |||
| Another study shows that the northern and southern Han Chinese are genetically close to each other and it finds that the genetic characteristics of present-day northern Han Chinese were already formed prior to three thousand years ago in the ].<ref name=":0" /> | |||
| A recent genetic study on the remains of people (~4,000 years ]) from the Mogou site in the ]-] (or Ganqing) region of China revealed more information on the genetic contributions of these ancient ] people to the ] of the Northern Han. It was deduced that 3,300 to 3,800 years ago some Mogou people had merged into the ancestral Han population, resulting in the Mogou people being similar to some northern Han in sharing up to ~33% paternal (O3a) and ~70% maternal (D, A, F, M10) haplogroups. The mixing ratio was possibly 13–18%.<ref>{{Cite journal |vauthors=Li J, Zeng W, Zhang Y, Ko AM, Li C, Zhu H, Fu Q, Zhou H |date=December 2017 |title=Ancient DNA reveals genetic connections between early Di-Qiang and Han Chinese |journal=BMC Evolutionary Biology |volume=17 |issue=1 |pages=239 |doi=10.1186/s12862-017-1082-0 |pmc=5716020 |pmid=29202706 |bibcode=2017BMCEE..17..239L |doi-access=free}}</ref> | |||
| A 2018 study calculated pairwise F<sub>ST</sub> (a measure of genetic difference) based on genome-wide SNPs, among the Han Chinese (Northern Han from ] and Southern Han from ], ] and ] provinces), Japanese and Korean populations sampled. It found that the smallest F<sub>ST</sub> value was between Northern Han Chinese (Beijing) (CHB) and Southern Han (Hunan, Fujian, etc.) Chinese (CHS) (F<sub>ST</sub> = 0.0014), while CHB and Korean (KOR) (F<sub>ST</sub> = 0.0026) and between KOR and Japanese (JPT) (F<sub>ST</sub> = 0.0033). Generally, pairwise F<sub>ST</sub> between Han Chinese, Japanese and Korean (0.0026~ 0.0090) are greater than that within Han Chinese (0.0014). These results suggested Han Chinese, Japanese and Korean are different in terms of genetic make-up, and the differences among the three groups are much larger than that between northern and southern Han Chinese.<ref name="Wang_2018">{{cite journal |vauthors=Wang Y, Lu D, Chung YJ, Xu S |title=Genetic structure, divergence and admixture of Han Chinese, Japanese and Korean populations |journal=Hereditas |volume=155 |issue=1 |pages=19 |date=6 April 2018 |pmid=29636655 |pmc=5889524 |doi=10.1186/s41065-018-0057-5 |doi-access=free }} | |||
| *{{cite web | vauthors = Xu S |date=10 April 2018 |title=Common ancestor of Han Chinese, Japanese and Koreans dated to 3000 – 3600 years ago |url=http://blogs.biomedcentral.com/on-biology/2018/04/10/common-ancestor-of-han-chinese-japanese-and-koreans-dated-to-3000-3600-years-ago/ |website=Biomed Central}}</ref> Nonetheless, there is also genetic diversity among the Southern Han Chinese. The genetic composition of the Han population in Fujian might not accurately represent that of the Han population in Guangdong. | |||
| The estimated contribution of northern Han to southern Han is substantial in both paternal and maternal lineages and a geographic ] exists for mtDNA. As a result, the northern Han are one of the primary contributors to the gene pool of the southern Han. However, it is noteworthy that the expansion process was not only dominated by males, as is shown by both contribution of the Y-chromosome and the mtDNA from northern Han to southern Han. Northern Han Chinese and Southern Han Chinese exhibit both Ancient Northern East Asian and Ancient Southern East Asian ancestries.<ref name=":6" /> These genetic observations are in line with historical records of continuous and large migratory waves of northern China inhabitants escaping warfare and famine, to southern China. Aside from these large migratory waves, other smaller southward migrations occurred during almost all periods in the past two millennia.<ref name="Wen2004" /> A study by the ] into the gene frequency data of Han subpopulations and ethnic minorities in China showed that Han subpopulations in different regions are also genetically quite close to the local ethnic minorities, suggesting that in many cases, ethnic minorities ancestry had mixed with Han, while at the same time, the Han ancestry had also mixed with the local ethnic minorities.<ref>{{cite journal |vauthors=Du R, Xiao C, Cavalli-Sforza LL |date=December 1997 |title=Genetic distances between Chinese populations calculated on gene frequencies of 38 loci |journal=Science in China Series C: Life Sciences |volume=40 |issue=6 |pages=613–21 |doi=10.1007/BF02882691 |pmid=18726285 |s2cid=1924085}}</ref> | |||
| ]) populations, and ] (KHV). The Chinese Han population could be mainly distinguished into 7 population clusters, including Northwest Han (], ]), North Han (Beijing, ], ], ], ], ], ], ] and ]), East Han (Jiangsu, ], ] and ]), Central Han (Hubei), Southeast Han (Fujian), South Han (], ], ], Hunan, ], ]) and ] Han (], ]) <ref>{{cite journal | vauthors = Cao Y, Li L, Xu M, Feng Z, Sun X, Lu J, Xu Y, Du P, Wang T, Hu R, Ye Z, Shi L, Tang X, Yan L, Gao Z, Chen G, Zhang Y, Chen L, Ning G, Bi Y, Wang W | display-authors = 6 | title = The ChinaMAP analytics of deep whole genome sequences in 10,588 individuals | journal = Cell Research | volume = 30 | issue = 9 | pages = 717–731 | date = September 2020 | pmid = 32355288 | pmc = 7609296 | doi = 10.1038/s41422-020-0322-9 }}</ref>]] | |||
| Han Chinese, similar to other East Asian populations, have inherited West Eurasian ancestry, around 2.8% in Northern Han Chinese and around 1.7% in Southern Han Chinese.<ref>{{Cite journal |last1=Qin |first1=Pengfei |last2=Zhou |first2=Ying |last3=Lou |first3=Haiyi |last4=Lu |first4=Dongsheng |last5=Yang |first5=Xiong |last6=Wang |first6=Yuchen |last7=Jin |first7=Li |last8=Chung |first8=Yeun-Jun |last9=Xu |first9=Shuhua |date=2 April 2015 |title=Quantitating and Dating Recent Gene Flow between European and East Asian Populations |journal=Scientific Reports |language=en |volume=5 |issue=1 |pages=9500 |doi=10.1038/srep09500 |pmid=25833680 |issn=2045-2322|pmc=4382708|bibcode=2015NatSR...5.9500Q }}</ref> | |||
| Another study shows that the northern and southern Han Chinese are genetically close to each other and it finds that the genetic characteristics of present-day northern Han Chinese were already formed prior to three thousand years ago in the ].<ref name=":0">{{Cite journal |vauthors=Zhao YB, Zhang Y, Zhang QC, Li HJ, Cui YQ, Xu Z, Jin L, Zhou H, Zhu H |date=2015 |title=Ancient DNA reveals that the genetic structure of the northern Han Chinese was shaped prior to 3,000 years ago |journal=PLOS ONE |volume=10 |issue=5 |pages=e0125676 |bibcode=2015PLoSO..1025676Z |doi=10.1371/journal.pone.0125676 |pmc=4418768 |pmid=25938511 |doi-access=free}}</ref> | |||
| An extensive, genome-wide association study of the Han population in 2008, shows that geographic-genetic stratification from north to south has occurred and centrally placed populations act as the conduit for outlying ones.<ref>{{cite journal | vauthors = Chen J, Zheng H, Bei JX, Sun L, Jia WH, Li T, Zhang F, Seielstad M, Zeng YX, Zhang X, Liu J | display-authors = 6 | title = Genetic structure of the Han Chinese population revealed by genome-wide SNP variation | journal = American Journal of Human Genetics | volume = 85 | issue = 6 | pages = 775–785 | date = December 2009 | pmid = 19944401 | pmc = 2790583 | doi = 10.1016/j.ajhg.2009.10.016}}</ref> Ultimately, with the exception in some ] branches of the Han Chinese, such as ], there is "coherent genetic structure" (homogeneity)<!-- "coherent genetic structure" doesn't seem to have a standard, formal definition--> in all Han Chinese.<ref name="Gan2008" /> | |||
| A recent genetic study on the remains of people (~4,000 years ]) from the Mogou site in the ]-] (or Ganqing) region of China revealed more information on the genetic contributions of these ancient ] people to the ] of the Northern Han. It was deduced that 3,300 to 3,800 years ago some Mogou people had merged into the ancestral Han population, resulting in the Mogou people being similar to some northern Han in sharing up to ~33% paternal (O3a) and ~70% maternal (D, A, F, M10) haplogroups. The mixture rate was possibly 13–18%.<ref>{{Cite journal |vauthors=Li J, Zeng W, Zhang Y, Ko AM, Li C, Zhu H, Fu Q, Zhou H |date=December 2017 |title=Ancient DNA reveals genetic connections between early Di-Qiang and Han Chinese |journal=BMC Evolutionary Biology |volume=17 |issue=1 |pages=239 |doi=10.1186/s12862-017-1082-0 |pmc=5716020 |pmid=29202706 |doi-access=free }}</ref> | |||
| ==== Paternal lineages ==== | |||
| ] | |||
| The major haplogroups of Han Chinese belong to subclades of ]. Y-chromosome ] is a common DNA marker in Han Chinese, as it appeared in China in prehistoric times, and is found in approximately 50% of Chinese males, with frequencies tending to be high toward the east of the country, ranging from 29.7% to 52% in Han from southern and central China, to 55–68% in Han from the eastern and northeastern Chinese mainland and Taiwan.<ref name=":3" /> | |||
| Other Y-DNA haplogroups that have been found with notable frequency in samples of Han Chinese include ] (9.1–13.0%), ] (6.0–12.0%), ] (3.6–10.3%), ] (4.7–7%), and ] (2/168 = 1.2–4.2%).<ref name="Lu2009" /><ref name=":3" /> | |||
| The estimated contribution of northern Han to southern Han is substantial in both paternal and maternal lineages and a geographic ] exists for mtDNA. As a result, the northern Han are one of the primary contributors to the gene pool of the southern Han. However, it is noteworthy that the expansion process was not only dominated by males, as is shown by both contribution of the Y-chromosome and the mtDNA from northern Han to southern Han. Northern Han Chinese and Southern Han Chinese exhibit both Ancient Northern East Asian and Ancient Southern East Asian ancestries.<ref name=":6" /> These genetic observations are in line with historical records of continuous and large migratory waves of northern China inhabitants escaping warfare and famine, to southern China. Aside from these large migratory waves, other smaller southward migrations occurred during almost all periods in the past two millennia.<ref name="Wen2004"/> A study by the ] into the gene frequency data of Han subpopulations and ethnic minorities in China showed that Han subpopulations in different regions are also genetically quite close to the local ethnic minorities, suggesting that in many cases, ethnic minorities ancestry had mixed with Han, while at the same time, the Han ancestry had also mixed with the local ethnic minorities.<ref>{{cite journal |vauthors=Du R, Xiao C, Cavalli-Sforza LL |date=December 1997 |title=Genetic distances between Chinese populations calculated on gene frequencies of 38 loci |journal=Science in China Series C: Life Sciences |volume=40 |issue=6 |pages=613–21 |doi=10.1007/BF02882691 |pmid=18726285 |s2cid=1924085}}</ref> | |||
| ==== Maternal lineages ==== | |||
| An extensive, genome-wide association study of the Han population in 2008, shows that geographic-genetic stratification from north to south has occurred and centrally placed populations act as the conduit for outlying ones.<ref>{{cite journal | vauthors = Chen J, Zheng H, Bei JX, Sun L, Jia WH, Li T, Zhang F, Seielstad M, Zeng YX, Zhang X, Liu J | display-authors = 6 | title = Genetic structure of the Han Chinese population revealed by genome-wide SNP variation | journal = American Journal of Human Genetics | volume = 85 | issue = 6 | pages = 775–785 | date = December 2009 | pmid = 19944401 | pmc = 2790583 | doi = 10.1016/j.ajhg.2009.10.016 }}</ref> Ultimately, with the exception in some ] branches of the Han Chinese, such as ], there is "coherent genetic structure" (homogeneity)<!-- "coherent genetic structure" doesn't seem to have a standard, formal definition--> in all Han Chinese.<ref name="Gan2008" /> | |||
| The mitochondrial-DNA haplogroups of the Han Chinese can be classified into the northern East Asian-dominating haplogroups, including A, C, D, G, M8, M9, and Z, and the southern East Asian-dominating haplogroups, including B, F, M7, N*, and R.<ref name="Wen2004" /> | |||
| === Paternal lineages === | |||
| Y-chromosome ] is a common DNA marker in Han Chinese, as it appeared in China in prehistoric times. It is found in more than 50% of Chinese males, with frequencies tending to be high toward the east of the country, ranging from 29.7% to 52% in Han from southern and central China,<ref name="Gan2008" /><ref name="Hammer2006" /><ref name="Xue2006" /><ref name="Kim2011" /><ref name="Yin2020">{{cite journal |vauthors=Yin C, Su K, He Z, Zhai D, Guo K, Chen X, Jin L, Li S |date=July 2020 |title=Genetic Reconstruction and Forensic Analysis of Chinese Shandong and Yunnan Han Populations by Co-Analyzing Y Chromosomal STRs and SNPs |journal=Genes |volume=11 |issue=7 |page=743 |doi=10.3390/genes11070743 |pmc=7397191 |pmid=32635262 |doi-access=free}}</ref><ref name="Xue2006" /><ref name="Yao2021">{{cite journal | vauthors = Yao H, Wang M, Zou X, Li Y, Yang X, Li A, Yeh HY, Wang P, Wang Z, Bai J, Guo J, Chen J, Ding X, Zhang Y, Lin B, Wang CC, He G | display-authors = 6 | title = New insights into the fine-scale history of western-eastern admixture of the northwestern Chinese population in the Hexi Corridor via genome-wide genetic legacy | journal = Molecular Genetics and Genomics | volume = 296 | issue = 3 | pages = 631–651 | date = May 2021 | pmid = 33650010 | doi = 10.1007/s00438-021-01767-0 }}</ref><ref name="pmid33650010">{{Cite journal |vauthors=Yao H, Wang M, Zou X, Li Y, Yang X, Li A, Yeh HY, Wang P, Wang Z, Bai J, Guo J, Chen J, Ding X, Zhang Y, Lin B, Wang CC, He G |date=May 2021 |title=New insights into the fine-scale history of western-eastern admixture of the northwestern Chinese population in the Hexi Corridor via genome-wide genetic legacy |journal=Molecular Genetics and Genomics |volume=296 |issue=3 |pages=631–651 |doi=10.1007/s00438-021-01767-0 |pmid=33650010 |s2cid=232091731}}</ref><ref name="Yan2011" /><ref name="Xue2006" /><ref name="Su1999">{{Cite journal |vauthors=Su B, Xiao J, Underhill P, Deka R, Zhang W, Akey J, Huang W, Shen D, Lu D, Luo J, Chu J, Tan J, Shen P, Davis R, Cavalli-Sforza L, Chakraborty R, Xiong M, Du R, Oefner P, Chen Z, Jin L |date=December 1999 |title=Y-Chromosome evidence for a northward migration of modern humans into Eastern Asia during the last Ice Age |journal=American Journal of Human Genetics |volume=65 |issue=6 |pages=1718–24 |doi=10.1086/302680 |pmc=1288383 |pmid=10577926}}</ref> to 55-68% in Han from the eastern and northeastern Chinese mainland and Taiwan.<ref name="Su1999"/><ref name="Tajima2004"/><ref name="Yan2011" /><ref name="Hammer2006" /><ref name="Trejaut2014"/><ref name="Yan2011" /><ref name="Trejaut2014">{{Cite journal |vauthors=Trejaut JA, Poloni ES, Yen JC, Lai YH, Loo JH, Lee CL, He CL, Lin M |date=June 2014 |title=Taiwan Y-chromosomal DNA variation and its relationship with Island Southeast Asia |journal=BMC Genetics |volume=15 |pages=77 |doi=10.1186/1471-2156-15-77 |pmc=4083334 |pmid=24965575 |doi-access=free }}</ref><ref name="Trejaut2014"/><ref name="Tajima2004"/><ref name="Yin2020"/><ref name="Xue2006" /><ref name="Hammer2006" /><ref>{{cite journal |display-authors=6 |vauthors=Yan S, Wang CC, Zheng HX, Wang W, Qin ZD, Wei LH, Wang Y, Pan XD, Fu WQ, He YG, Xiong LJ, Jin WF, Li SL, An Y, Li H, Jin L |year=2014 |title=Y chromosomes of 40% Chinese descend from three Neolithic super-grandfathers |journal=PLOS ONE |volume=9 |issue=8 |pages=e105691 |arxiv=1310.3897 |bibcode=2014PLoSO...9j5691Y |doi=10.1371/journal.pone.0105691 |pmc=4149484 |pmid=25170956 |doi-access=free}}</ref><ref name="Hurles2005">{{Cite journal |vauthors=Hurles ME, Sykes BC, Jobling MA, Forster P |date=May 2005 |title=The dual origin of the Malagasy in Island Southeast Asia and East Africa: evidence from maternal and paternal lineages |journal=American Journal of Human Genetics |volume=76 |issue=5 |pages=894–901 |doi=10.1086/430051 |pmc=1199379 |pmid=15793703}}</ref> | |||
| Other Y-DNA haplogroups that have been found with notable frequency in samples of Han Chinese include ] (9.1%-13.0%), ] (6.0%-12.0%), ] (3.6%-10.3%), ] (4.7-7%<ref name="Lu2009">{{Cite journal |display-authors=6 |vauthors=Lu C, Zhang J, Li Y, Xia Y, Zhang F, Wu B, Wu W, Ji G, Gu A, Wang S, Jin L, Wang X |date=March 2009 |title=The b2/b3 subdeletion shows higher risk of spermatogenic failure and higher frequency of complete AZFc deletion than the gr/gr subdeletion in a Chinese population |journal=Human Molecular Genetics |volume=18 |issue=6 |pages=1122–1130 |doi=10.1093/hmg/ddn427 |pmid=19088127 |doi-access=free}}</ref>), and ] (2/168 = 1.2-4.2%<ref name="Lu2009"/>). | |||
| However, the ] (mtDNA) of Han Chinese increases in diversity as one looks from northern to southern China, which suggests that male migrants from northern China married with women from local peoples after arriving in modern-day Guangdong, Shanghai, Nanjing, Fujian, and other regions of southern China.<ref name="Wen2004">{{Cite journal |display-authors=6 |vauthors=Wen B, Li H, Lu D, Song X, Zhang F, He Y, Li F, Gao Y, Mao X, Zhang L, Qian J, Tan J, Jin J, Huang W, Deka R, Su B, Chakraborty R, Jin L |date=September 2004 |title=Genetic evidence supports demic diffusion of Han culture |journal=Nature |volume=431 |issue=7006 |pages=302–305 |bibcode=2004Natur.431..302W |doi=10.1038/nature02878 |pmid=15372031 |s2cid=4301581}}</ref><ref name="EJH">{{Cite journal |display-authors=6 |vauthors=Xue F, Wang Y, Xu S, Zhang F, Wen B, Wu X, Lu M, Deka R, Qian J, Jin L |date=June 2008 |title=A spatial analysis of genetic structure of human populations in China reveals distinct difference between maternal and paternal lineages |journal=European Journal of Human Genetics |volume=16 |issue=6 |pages=705–717 |doi=10.1038/sj.ejhg.5201998 |pmid=18212820 |doi-access=free}}</ref> | |||
| However, a new study in 2019, shows that ] (mtDNA) doesn't increase in diversity from northern to southern China. Most of the mtDNA haplogroups carried by southern Han Chinese are also carried by northern Han Chinese.<ref>{{cite journal | vauthors = Li YC, Ye WJ, Jiang CG, Zeng Z, Tian JY, Yang LQ, Liu KJ, Kong QP | display-authors = 6 | title = River Valleys Shaped the Maternal Genetic Landscape of Han Chinese | journal = Molecular Biology and Evolution | volume = 36 | issue = 8 | pages = 1643–1652 | date = August 2019 | pmid = 31112995 | url = https://academic.oup.com/mbe/article/36/8/1643/5423190 }}</ref> | |||
| === Maternal lineages === | |||
| The mitochondrial-DNA haplogroups of the Han Chinese can be classified into the northern East Asian-dominating haplogroups, including A, C, D, G, M8, M9, and Z, and the southern East Asian-dominating haplogroups, including B, F, M7, N*, and R.<ref name="Wen2004"/> | |||
| These haplogroups account for 52.7% and 33.85% of those in the Northern Han, respectively. Haplogroup D is the modal mtDNA haplogroup among northern East Asians. Among these haplogroups, D, B, F, and A were predominant in the Northern Han, with frequencies of 25.77%, 11.54%, 11.54%, and 8.08%, respectively. | These haplogroups account for 52.7% and 33.85% of those in the Northern Han, respectively. Haplogroup D is the modal mtDNA haplogroup among northern East Asians. Among these haplogroups, D, B, F, and A were predominant in the Northern Han, with frequencies of 25.77%, 11.54%, 11.54%, and 8.08%, respectively. | ||
| However, in the Southern Han, the northern and southern East Asian-dominating mtDNA haplogroups accounted for 35.62% and 51.91%, respectively. The frequencies of haplogroups D, B, F, and A reached 15.68%, 20.85%, 16.29%, and 5.63%, respectively.<ref name=":0"/><ref>{{Cite journal |vauthors=Yao YG, Kong QP, Bandelt HJ, Kivisild T, Zhang YP |date=March 2002 |title=Phylogeographic differentiation of mitochondrial DNA in Han Chinese |journal=American Journal of Human Genetics |volume=70 |issue=3 |pages=635–51 |doi=10.1086/338999 |pmc=384943 |pmid=11836649}}</ref><ref>{{Cite journal |vauthors=Kivisild T, Tolk HV, Parik J, Wang Y, Papiha SS, Bandelt HJ, Villems R |date=October 2002 |title=The emerging limbs and twigs of the East Asian mtDNA tree |journal=Molecular Biology and Evolution |volume=19 |issue=10 |pages=1737–51 |doi=10.1093/oxfordjournals.molbev.a003996 |pmid=12270900 |doi-access=free}}</ref><ref>{{Cite journal |vauthors=Yao YG, Kong QP, Man XY, Bandelt HJ, Zhang YP |date=February 2003 |title=Reconstructing the evolutionary history of China: a caveat about inferences drawn from ancient DNA |journal=Molecular Biology and Evolution |volume=20 |issue=2 |pages=214–9 |doi=10.1093/molbev/msg026 |pmid=12598688 |doi-access=free}}</ref><ref>{{Cite journal |vauthors=Kong QP, Sun C, Wang HW, Zhao M, Wang WZ, Zhong L, Hao XD, Pan H, Wang SY, Cheng YT, Zhu CL, Wu SF, Liu LN, Jin JQ, Yao YG, Zhang YP |date=January 2011 |title=Large-scale mtDNA screening reveals a surprising matrilineal complexity in east Asia and its implications to the peopling of the region |journal=Molecular Biology and Evolution |volume=28 |issue=1 |pages=513–22 |doi=10.1093/molbev/msq219 |pmid=20713468 |doi-access=free}}</ref> | However, in the Southern Han, the northern and southern East Asian-dominating mtDNA haplogroups accounted for 35.62% and 51.91%, respectively. The frequencies of haplogroups D, B, F, and A reached 15.68%, 20.85%, 16.29%, and 5.63%, respectively.<ref name=":0" /><ref>{{Cite journal |vauthors=Yao YG, Kong QP, Bandelt HJ, Kivisild T, Zhang YP |date=March 2002 |title=Phylogeographic differentiation of mitochondrial DNA in Han Chinese |journal=American Journal of Human Genetics |volume=70 |issue=3 |pages=635–51 |doi=10.1086/338999 |pmc=384943 |pmid=11836649}}</ref><ref>{{Cite journal |vauthors=Kivisild T, Tolk HV, Parik J, Wang Y, Papiha SS, Bandelt HJ, Villems R |date=October 2002 |title=The emerging limbs and twigs of the East Asian mtDNA tree |journal=Molecular Biology and Evolution |volume=19 |issue=10 |pages=1737–51 |doi=10.1093/oxfordjournals.molbev.a003996 |pmid=12270900 |doi-access=free}}</ref><ref>{{Cite journal |vauthors=Yao YG, Kong QP, Man XY, Bandelt HJ, Zhang YP |date=February 2003 |title=Reconstructing the evolutionary history of China: a caveat about inferences drawn from ancient DNA |journal=Molecular Biology and Evolution |volume=20 |issue=2 |pages=214–9 |doi=10.1093/molbev/msg026 |pmid=12598688 |doi-access=free}}</ref><ref>{{Cite journal |vauthors=Kong QP, Sun C, Wang HW, Zhao M, Wang WZ, Zhong L, Hao XD, Pan H, Wang SY, Cheng YT, Zhu CL, Wu SF, Liu LN, Jin JQ, Yao YG, Zhang YP |date=January 2011 |title=Large-scale mtDNA screening reveals a surprising matrilineal complexity in east Asia and its implications to the peopling of the region |journal=Molecular Biology and Evolution |volume=28 |issue=1 |pages=513–22 |doi=10.1093/molbev/msq219 |pmid=20713468 |doi-access=free}}</ref> | ||
| === Tibetan peoples === | |||
| == Genetic history of Tibetans == | |||
| {{ |
{{Main|Tibetan people}} | ||
| The ethnic roots of Tibetans can be traced back to a deep Eastern Asian lineage representing the indigenous population of the Tibetan plateau since c. 40,000 to 30,000 years ago, and arriving Neolithic farmers from the ] within the last 10,000 years associated, and which can be associated with having introduced the ]. Modern Tibetans derive up to 20% from Paleolithic Tibetans, with the remaining 80% being primarily derived from Yellow River farmers. The present-day Tibetan gene pool was formed at least 5,100 years BP.<ref>{{Cite journal |last1=Liu |first1=Chi-Chun |last2=Witonsky |first2=David |last3=Gosling |first3=Anna |last4=Lee |first4=Ju Hyeon |last5=Ringbauer |first5=Harald |last6=Hagan |first6=Richard |last7=Patel |first7=Nisha |last8=Stahl |first8=Raphaela |last9=Novembre |first9=John |last10=Aldenderfer |first10=Mark |last11=Warinner |first11=Christina |last12=Di Rienzo |first12=Anna |last13=Jeong |first13=Choongwon |date=8 March 2022 |title=Ancient genomes from the Himalayas illuminate the genetic history of Tibetans and their Tibeto-Burman speaking neighbors |journal=Nature Communications |language=en |volume=13 |issue=1 |pages=1203 |bibcode=2022NatCo..13.1203L |doi=10.1038/s41467-022-28827-2 |issn=2041-1723 |pmid=35260549 |pmc=8904508 |s2cid=247317520}}</ref><ref>{{Cite journal |last1=Wang |first1=Hongru |last2=Yang |first2=Melinda A. |last3=Wangdue |first3=Shargan |last4=Lu |first4=Hongliang |last5=Chen |first5=Honghai |last6=Li |first6=Linhui |last7=Dong |first7=Guanghui |last8=Tsring |first8=Tinley |last9=Yuan |first9=Haibing |last10=He |first10=Wei |last11=Ding |first11=Manyu |last12=Wu |first12=Xiaohong |last13=Li |first13=Shuai |last14=Tashi |first14=Norbu |last15=Yang |first15=Tsho |date=15 March 2023 |title=Human genetic history on the Tibetan Plateau in the past 5100 years |journal=Science Advances |language=en |volume=9 |issue=11 |pages=eadd5582 |bibcode=2023SciA....9D5582W |doi=10.1126/sciadv.add5582 |issn=2375-2548 |pmc=10022901 |pmid=36930720}}</ref> | |||
| Modern Tibetan populations are genetically most similar to other modern ] populations.<ref name="Lu_2016"/> They also show more genetic affinity for modern ] than modern Siberian populations.<ref name="Lu_2016"/> | |||
| ==== Paternal lineage ==== | |||
| A 2016 study found that the Tibetan gene pool diverged from that of ] around 15,000 years ago, which can be largely attributed to post-LGM (]) arrivals. Analysis of around 200 contemporary populations showed that Tibetans share ancestry with populations from East Asia (~82%), Central Asia and Siberia (~11%), South Asia (~6%), and western Eurasia and Oceania (~1%). These results support that Tibetans arose from a mixture of multiple ancestral gene pools but that their origins are much more complicated and ancient than previously suspected.<ref name="Lu_2016">{{Cite journal |display-authors=6 |vauthors=Lu D, Lou H, Yuan K, Wang X, Wang Y, Zhang C, Lu Y, Yang X, Deng L, Zhou Y, Feng Q, Hu Y, Ding Q, Yang Y, Li S, Jin L, Guan Y, Su B, Kang L, Xu S |date=September 2016 |title=Ancestral Origins and Genetic History of Tibetan Highlanders |journal=American Journal of Human Genetics |volume=99 |issue=3 |pages=580–594 |doi=10.1016/j.ajhg.2016.07.002 |pmc=5011065 |pmid=27569548}}</ref> | |||
| Tibetan males predominantly belong to the paternal lineage ] followed by lower amounts of ].<ref>{{cite journal |last1=Bhandari |first1=Sushil |last2=Zhang |first2=Xiaoming |date=5 November 2015 |title=Genetic evidence of a recent Tibetan ancestry to Sherpas in the Himalayan region |journal=Scientific Reports |volume=5 |pages=16249 |bibcode=2015NatSR...516249B |doi=10.1038/srep16249 |issn=2045-2322 |pmc=4633682 |pmid=26538459}} "Comparing Sherpas, Tibetans, and Han Chinese showed that the D-M174 is the predominant haplogroup in Sherpas (43.38%) and prevalent in Tibetans (52.84%)5, but rare among both Han Chinese (1.4–6.51%)6,7 and other Asian populations (0.02–0.07%)8, aside from Japanese (34.7%) who possesses a distinct D-M174 lineage highly diverged from those in Tibetans and other Asian populations9,10."</ref> | |||
| === |
==== Maternal lineage ==== | ||
| Tibetan females belong mainly to the Northeast Asian maternal haplogroups M9a1a, M9a1b, D4g2, D4i and G2ac, showing continuity with ancient middle and upper ] populations.<ref>{{cite journal |last1=Zhang |first1=Ganyu |last2=Cui |first2=Can |last3=Wangdue |first3=Shargan |date=16 March 2023 |title=Maternal genetic history of ancient Tibetans over the past 4000 years |journal=Journal of Genetics and Genomics |language=en |volume=50 |issue=10 |pages=765–775 |doi=10.1016/j.jgg.2023.03.007 |pmid=36933795 |s2cid=257588399|doi-access=free}}</ref> | |||
| A study in 2010 suggested that the majority of the Tibetan gene pool may have diverged from the Zang around 15,000 years ago.<ref>{{Cite journal |display-authors=6 |vauthors=Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZX, Pool JE, Xu X, Jiang H, Vinckenbosch N, Korneliussen TS, Zheng H, Liu T, He W, Li K, Luo R, Nie X, Wu H, Zhao M, Cao H, Zou J, Shan Y, Li S, Yang Q, Ni P, Tian G, Xu J, Liu X, Jiang T, Wu R, Zhou G, Tang M, Qin J, Wang T, Feng S, Li G, Luosang J, Wang W, Chen F, Wang Y, Zheng X, Li Z, Bianba Z, Yang G, Wang X, Tang S, Gao G, Chen Y, Luo Z, Gusang L, Cao Z, Zhang Q, Ouyang W, Ren X, Liang H, Zheng H, Huang Y, Li J, Bolund L, Kristiansen K, Li Y, Zhang Y, Zhang X, Li R, Li S, Yang H, Nielsen R, Wang J, Wang J |date=July 2010 |title=Sequencing of 50 human exomes reveals adaptation to high altitude |journal=Science |volume=329 |issue=5987 |pages=75–8 |bibcode=2010Sci...329...75Y |doi=10.1126/science.1190371 |pmc=3711608 |pmid=20595611 }}</ref> However, there are possibilities of much earlier human inhabitation of Tibet,<ref name="Aldenderfer">{{Cite journal |vauthors=Aldenderfer M, Yinong Z |date=March 2004 |title=The Prehistory of the Tibetan Plateau to the Seventh Century A.D.: Perspectives and Research from China and the West Since 1950 |journal=Journal of World Prehistory |volume=18 |issue=1 |pages=1–55 |doi=10.1023/B:JOWO.0000038657.79035.9e |s2cid=154022638}}</ref><ref name="Yuan">{{Cite journal |vauthors=Yuan B, Huang W, Zhang D |date=October 2007 |title=New evidence for human occupation of the northern Tibetan Plateau, China during the Late Pleistocene |journal=Chinese Science Bulletin |volume=52 |issue=19 |pages=2675–2679 |bibcode=2007ChSBu..52.2675Y |doi=10.1007/s11434-007-0357-z |s2cid=93228078}}</ref> and these early residents may have contributed to the modern Tibetan gene pool.<ref name="Zhao">{{Cite journal |display-authors=6 |vauthors=Zhao M, Kong QP, Wang HW, Peng MS, Xie XD, Wang WZ, Duan JG, Cai MC, Zhao SN, Tu YQ, Wu SF, Yao YG, Bandelt HJ, Zhang YP |date=December 2009 |title=Mitochondrial genome evidence reveals successful Late Paleolithic settlement on the Tibetan Plateau |journal=Proceedings of the National Academy of Sciences of the United States of America |volume=106 |issue=50 |pages=21230–5 |bibcode=2009PNAS..10621230Z |doi=10.1073/pnas.0907844106 |pmc=2795552 |pmid=19955425 |doi-access=free }}</ref> | |||
| === Turkic peoples === | |||
| The date of divergence between Tibetans and Sherpas was estimated to have taken place around 11,000 to 7,000 years ago.<ref name="Lu_2016"/> | |||
| {{See also|Turkic peoples}} | |||
| Linguistic and genetic evidence strongly suggests an early presence of Turkic peoples in eastern Mongolia.<ref>{{Cite book | vauthors = Blench R, Spriggs M |url=https://books.google.com/books?id=48iKiprsRMwC |title=Archaeology and Language II: Archaeological Data and Linguistic Hypotheses |date=2 September 2003 |publisher=Routledge |isbn=978-1-134-82869-2 |language=en}}</ref> The genetic evidence suggests that the ] of Central Asia was carried out by ] ] migrating out of Mongolia.<ref name=":5" /> | |||
| === Relationship to archaic hominins === | |||
| After modern Oceanic populations, modern Tibetan populations show the highest rate of allele sharing with archaic hominins at over 6%.<ref name="Lu_2016"/> Modern Tibetans show genetic affinities to three archaic populations: ], ], and an unidentified archaic population.<ref name="Lu_2016"/> | |||
| Genetic data found that almost all modern Turkic-speaking peoples retained at least some shared ancestry associated with "Southern Siberian and Mongolian" (SSM) populations, supporting this region as the "Inner Asian Homeland (IAH) of the pioneer carriers of Turkic languages" which subsequently expanded into Central Asia.<ref>{{Cite journal |last1=Yunusbayev |first1=Bayazit |last2=Metspalu |first2=Mait |last3=Metspalu |first3=Ene |last4=Valeev |first4=Albert |last5=Litvinov |first5=Sergei |last6=Valiev |first6=Ruslan |last7=Akhmetova |first7=Vita |last8=Balanovska |first8=Elena |last9=Balanovsky |first9=Oleg |last10=Turdikulova |first10=Shahlo |last11=Dalimova |first11=Dilbar |last12=Nymadawa |first12=Pagbajabyn |last13=Bahmanimehr |first13=Ardeshir |last14=Sahakyan |first14=Hovhannes |last15=Tambets |first15=Kristiina |date=21 April 2015 |title=The Genetic Legacy of the Expansion of Turkic-Speaking Nomads across Eurasia |journal=PLOS Genetics |language=en |volume=11 |issue=4 |pages=e1005068 |doi=10.1371/journal.pgen.1005068 |issn=1553-7404 |pmc=4405460 |pmid=25898006 |doi-access=free}}</ref>] | |||
| In comparison to modern Han populations, modern Tibetans show greater genetic affinity to Denisovans; however, both the Han and Tibetans have similar ratios of genetic affinity to general Neanderthal populations.<ref name="Lu_2016"/> | |||
| Modern Tibetans were identified as the modern population that has the most alleles in common with ].<ref name="Lu_2016"/> | |||
| === Paternal lineage === | |||
| The distribution of ] (subclade ]) is found among nearly all the populations of Central Asia and Northeast Asia south of the Russian border, although generally at a low frequency of 2% or less. A dramatic spike in the frequency of D-M174 occurs as one approaches the ]. D-M174 is also found at high frequencies among ], but it fades into low frequencies in ] and ] between ] and Tibet. The claim that the ] and Tibetans are related, while ] since ], has not found support in genetic studies. Some light has been shed on their origins, however, by one genetic study in which it was indicated that Tibetan Y-chromosomes had multiple origins, one from Central Asia and the other from East Asia.<ref name="pmid11153912">{{Cite journal |vauthors=Su B, Xiao C, Deka R, Seielstad MT, Kangwanpong D, Xiao J, Lu D, Underhill P, Cavalli-Sforza L, Chakraborty R, Jin L |date=December 2000 |title=Y chromosome haplotypes reveal prehistorical migrations to the Himalayas |journal=Human Genetics |volume=107 |issue=6 |pages=582–90 |doi=10.1007/s004390000406 |pmid=11153912 |s2cid=36788262}}</ref> | |||
| == Genetic history of Turks == | |||
| {{See also|Turkic peoples|Genetic studies on Turkish people}} | |||
| The Turkic peoples are a collection of ]s of ], ], ] and ] as well as parts of ] and ]. They speak ] belonging to the ]. | |||
| Proposals for the homeland of the Turkic peoples and their language are far-ranging, from the ] to ] (]).<ref>{{Cite journal |display-authors=6 |vauthors=Yunusbayev B, Metspalu M, Metspalu E, Valeev A, Litvinov S, Valiev R, Akhmetova V, Balanovska E, Balanovsky O, Turdikulova S, Dalimova D, Nymadawa P, Bahmanimehr A, Sahakyan H, Tambets K, Fedorova S, Barashkov N, Khidiyatova I, Mihailov E, Khusainova R, Damba L, Derenko M, Malyarchuk B, Osipova L, Voevoda M, Yepiskoposyan L, Kivisild T, Khusnutdinova E, Villems R |date=April 2015 |title=The genetic legacy of the expansion of Turkic-speaking nomads across Eurasia |journal=PLOS Genetics |volume=11 |issue=4 |pages=e1005068 |doi=10.1371/journal.pgen.1005068 |pmc=4405460 |pmid=25898006 |quote=The origin and early dispersal history of the Turkic peoples is disputed, with candidates for their ancient homeland ranging from the Transcaspian steppe to Manchuria in Northeast Asia, |doi-access=free }}</ref> | |||
| According to Yunusbayev, genetic evidence points to an origin in the region near ] and ] as the "Inner Asian Homeland" of the Turkic ethnicity.<ref>{{Cite journal |display-authors=6 |vauthors=Yunusbayev B, Metspalu M, Metspalu E, Valeev A, Litvinov S, Valiev R, Akhmetova V, Balanovska E, Balanovsky O, Turdikulova S, Dalimova D, Nymadawa P, Bahmanimehr A, Sahakyan H, Tambets K, Fedorova S, Barashkov N, Khidiyatova I, Mihailov E, Khusainova R, Damba L, Derenko M, Malyarchuk B, Osipova L, Voevoda M, Yepiskoposyan L, Kivisild T, Khusnutdinova E, Villems R |date=April 2015 |title=The genetic legacy of the expansion of Turkic-speaking nomads across Eurasia |journal=PLOS Genetics |volume=11 |issue=4 |pages=e1005068 |doi=10.1371/journal.pgen.1005068 |pmc=4405460 |pmid=25898006 |quote="Thus, our study provides the first genetic evidence supporting one of the previously hypothesized IAHs to be near Mongolia and South Siberia." |doi-access=free }}</ref> | |||
| ] with Northeast Asian millet-agriculturalists, which later adopted a nomadic lifestyle and expanded from eastern Mongolia westwards.]] | ] with Northeast Asian millet-agriculturalists, which later adopted a nomadic lifestyle and expanded from eastern Mongolia westwards.]] | ||
| An ] origin of the early Turkic peoples has been corroborated in multiple recent studies. Early and medieval Turkic groups however exhibited a wide range of both (Northern) East Asian and West Eurasian genetic origins, in part through long-term contact with neighboring peoples such as Iranian, Mongolic, Tocharian, Uralic and Yeniseian peoples, and others.<ref>{{Cite journal |vauthors=Nelson S, Zhushchikhovskaya I, Li T, Hudson M, Robbeets M |date=2020 |title=Tracing population movements in ancient East Asia through the linguistics and archaeology of textile production |journal=Evolutionary Human Sciences |language=en |volume=2 |pages=e5 |doi=10.1017/ehs.2020.4 |pmid=37588355 |pmc=10427276 |issn=2513-843X |s2cid=213436897|doi-access=free}}</ref><ref>{{Cite journal |vauthors=Li T, Ning C, Zhushchikhovskaya IS, Hudson MJ, Robbeets M |date=1 June 2020 |title=Millet agriculture dispersed from Northeast China to the Russian Far East: Integrating archaeology, genetics, and linguistics |journal=Archaeological Research in Asia |language=en |volume=22 |pages=100177 |doi=10.1016/j.ara.2020.100177 |issn=2352-2267 |s2cid=213952845|doi-access=free |hdl=21.11116/0000-0005-D82B-8 |hdl-access=free}}</ref><ref>{{Cite journal |vauthors=Uchiyama J, Gillam JC, Savelyev A, Ning C |date=2020 |title=Populations dynamics in Northern Eurasian forests: a long-term perspective from Northeast Asia |journal=Evolutionary Human Sciences |language=en |volume=2 |pages=e16 |doi=10.1017/ehs.2020.11 |pmid=37588381 |pmc=10427466 |issn=2513-843X |s2cid=219470000|doi-access=free}}</ref><ref>{{Cite journal |last=Golden |first=Peter B. |date=October 2018 |title=The Ethnogonic Tales of the Türks |url=http://journals.sagepub.com/doi/10.1177/0971945818775373 |journal=The Medieval History Journal |language=en |volume=21 |issue=2 |pages=291–327 |doi=10.1177/0971945818775373 |s2cid=166026934 |issn=0971-9458}}</ref><ref>{{Cite journal |last1=Dai |first1=Shan-Shan |last2=Sulaiman |first2=Xierzhatijiang |last3=Isakova |first3=Jainagul |last4=Xu |first4=Wei-Fang |last5=Abdulloevich |first5=Najmudinov Tojiddin |last6=Afanasevna |first6=Manilova Elena |last7=Ibrohimovich |first7=Khudoidodov Behruz |last8=Chen |first8=Xi |last9=Yang |first9=Wei-Kang |last10=Wang |first10=Ming-Shan |last11=Shen |first11=Quan-Kuan |last12=Yang |first12=Xing-Yan |last13=Yao |first13=Yong-Gang |last14=Aldashev |first14=Almaz A |last15=Saidov |first15=Abdusattor |date=25 August 2022 |title=The Genetic Echo of the Tarim Mummies in Modern Central Asians |url=https://doi.org/10.1093/molbev/msac179 |journal=Molecular Biology and Evolution |volume=39 |issue=9 |doi=10.1093/molbev/msac179 |issn=0737-4038 |pmc=9469894 |pmid=36006373}}</ref><ref>{{Cite journal |last1=Guarino-Vignon |first1=Perle |last2=Marchi |first2=Nina |last3=Bendezu-Sarmiento |first3=Julio |last4=Heyer |first4=Evelyne |last5=Bon |first5=Céline |date=14 January 2022 |title=Genetic continuity of Indo-Iranian speakers since the Iron Age in southern Central Asia |journal=Scientific Reports |language=en |volume=12 |issue=1 |pages=733 |doi=10.1038/s41598-021-04144-4 |issn=2045-2322 |pmc=8760286 |pmid=35031610|bibcode=2022NatSR..12..733G}}</ref> | |||
| ==== Paternal lineages ==== | |||
| More recent genetic and archeologic studies suggest that the Turkic peoples were descended from a Transeurasian agricultural community based in northeast ], which is to be associated with the ] and the succeeding ].<ref>{{Cite journal |vauthors=Robbeets M |date=January 2020 |title=The Transeurasian homeland: where, what and when? |url=https://www.academia.edu/43039094 |journal=The Oxford Guide to the Transeurasian Languages |page=772 |doi=10.1093/oso/9780198804628.003.0045 |isbn=978-0-19-880462-8|hdl=21.11116/0000-0006-642A-B |hdl-access=free }}</ref> The East Asian agricultural origin of the Turkic peoples has been corroborated in multiple recent studies. Around 2,200 BC, due to the desertification of northeast China, the agricultural ancestors of the Turkic peoples probably migrated westwards into ], where they adopted a pastoral lifestyle.<ref>{{Cite journal |vauthors=Nelson S, Zhushchikhovskaya I, Li T, Hudson M, Robbeets M |date=2020 |title=Tracing population movements in ancient East Asia through the linguistics and archaeology of textile production |journal=Evolutionary Human Sciences |language=en |volume=2 |pages=e5 |doi=10.1017/ehs.2020.4 |pmid=37588355 |pmc=10427276 |issn=2513-843X |s2cid=213436897|doi-access=free }}</ref><ref>{{Cite journal |vauthors=Li T, Ning C, Zhushchikhovskaya IS, Hudson MJ, Robbeets M |date=1 June 2020 |title=Millet agriculture dispersed from Northeast China to the Russian Far East: Integrating archaeology, genetics, and linguistics |journal=Archaeological Research in Asia |language=en |volume=22 |pages=100177 |doi=10.1016/j.ara.2020.100177 |issn=2352-2267 |s2cid=213952845|doi-access=free }}</ref><ref name="Uchiyama">{{Cite journal |vauthors=Uchiyama J, Gillam JC, Savelyev A, Ning C |date=2020 |title=Populations dynamics in Northern Eurasian forests: a long-term perspective from Northeast Asia |journal=Evolutionary Human Sciences |language=en |volume=2 |pages=e16 |doi=10.1017/ehs.2020.11 |pmid=37588381 |pmc=10427466 |issn=2513-843X |s2cid=219470000|doi-access=free }}</ref> | |||
| Common Y-DNA haplogroups in Turkic peoples are ] (found with especially high frequency among Turkic peoples living in present-day ], especially among ], as Zabolotnie Tatars have one of the highest frequencies of this haplogroup, second only to ] ] ), ] (especially in ], and in particular, ], also in ] among ]), ] (especially in ] among the ], also quite frequent among ] and among ] and the Qangly ]), and ] (especially among Turkic peoples living in present-day ], the Naiman ] and ]). Some groups also have ] (notably frequent among the ], ], and ] of Southern Siberia, the ] of the Southern ] region of ], and the ] ]), ] (notably frequent among the ], ], ], ], ], ] and several other Turkic peoples living in present-day Russia), ] (especially frequent among ], ], and ]), and ] (especially among ], but also observed regularly with low frequency among ], ], ], and ]).<ref>{{Cite journal |vauthors=Zerjal T, Wells RS, Yuldasheva N, Ruzibakiev R, Tyler-Smith C |date=September 2002 |title=A genetic landscape reshaped by recent events: Y-chromosomal insights into central Asia |journal=American Journal of Human Genetics |language=en |volume=71 |issue=3 |pages=466–82 |doi=10.1086/342096 |pmc=419996 |pmid=12145751}}</ref><ref>{{Cite book |title=Genomics and Health in the Developing World |vauthors=Kumar D |date=11 May 2012 |publisher=Oxford University Press |isbn=978-0-19-970547-4 |location=Oxford, England |pages=1265–1267}}</ref> | |||
| Linguistic and genetic evidence strongly suggests an early presence of Turkic peoples in eastern Mongolia.<ref>{{Cite book | vauthors = Blench R, Spriggs M |url=https://books.google.com/books?id=48iKiprsRMwC |title=Archaeology and Language II: Archaeological Data and Linguistic Hypotheses |date=2 September 2003 |publisher=Routledge |isbn=978-1-134-82869-2 |language=en}}</ref><ref name="The genetic legacy of the expansion">{{Cite journal |display-authors=6 |vauthors=Yunusbayev B, Metspalu M, Metspalu E, Valeev A, Litvinov S, Valiev R, Akhmetova V, Balanovska E, Balanovsky O, Turdikulova S, Dalimova D, Nymadawa P, Bahmanimehr A, Sahakyan H, Tambets K, Fedorova S, Barashkov N, Khidiyatova I, Mihailov E, Khusainova R, Damba L, Derenko M, Malyarchuk B, Osipova L, Voevoda M, Yepiskoposyan L, Kivisild T, Khusnutdinova E, Villems R |date=April 2015 |title=The genetic legacy of the expansion of Turkic-speaking nomads across Eurasia |journal=PLOS Genetics |volume=11 |issue=4 |pages=e1005068 |doi=10.1371/journal.pgen.1005068 |pmc=4405460 |pmid=25898006 |doi-access=free }}</ref> Genetic studies have shown that the early Turkic peoples were of predominantly East Asian origins with medieval Turkic samples being more heterogeneous, and that Turkic culture and language was spread westwards through elite dominance.<ref name="Uchiyama"/><ref>{{Cite journal |vauthors=Lee JY, Kuang S |date=18 October 2017 |title=A Comparative Analysis of Chinese Historical Sources and y-dna Studies with Regard to the Early and Medieval Turkic Peoples |url=https://brill.com/view/journals/inas/19/2/article-p197_197.xml |journal=Inner Asia |volume=19 |issue=2 |pages=197–239 |doi=10.1163/22105018-12340089 |issn=2210-5018 |s2cid=165623743|doi-access=free }}</ref> The genetic evidence suggests that the ] of Central Asia was carried out by ] ] migrating out of Mongolia.<ref>{{Cite journal |display-authors=6 |vauthors=Damgaard PB, Marchi N, Rasmussen S, Peyrot M, Renaud G, Korneliussen T, Moreno-Mayar JV, Pedersen MW, Goldberg A, Usmanova E, Baimukhanov N, Loman V, Hedeager L, Pedersen AG, Nielsen K, Afanasiev G, Akmatov K, Aldashev A, Alpaslan A, Baimbetov G, Bazaliiskii VI, Beisenov A, Boldbaatar B, Boldgiv B, Dorzhu C, Ellingvag S, Erdenebaatar D, Dajani R, Dmitriev E, Evdokimov V, Frei KM, Gromov A, Goryachev A, Hakonarson H, Hegay T, Khachatryan Z, Khaskhanov R, Kitov E, Kolbina A, Kubatbek T, Kukushkin A, Kukushkin I, Lau N, Margaryan A, Merkyte I, Mertz IV, Mertz VK, Mijiddorj E, Moiyesev V, Mukhtarova G, Nurmukhanbetov B, Orozbekova Z, Panyushkina I, Pieta K, Smrčka V, Shevnina I, Logvin A, Sjögren KG, Štolcová T, Taravella AM, Tashbaeva K, Tkachev A, Tulegenov T, Voyakin D, Yepiskoposyan L, Undrakhbold S, Varfolomeev V, Weber A, Wilson Sayres MA, Kradin N, Allentoft ME, Orlando L, Nielsen R, Sikora M, Heyer E, Kristiansen K, Willerslev E |date=May 2018 |title=137 ancient human genomes from across the Eurasian steppes |journal=Nature |volume=557 |issue=7705 |pages=369–374 |bibcode=2018Natur.557..369D |doi=10.1038/s41586-018-0094-2 |pmid=29743675 |hdl=1887/3202709 |s2cid=13670282|hdl-access=free }}</ref><references group="note"/> | |||
| === Paternal lineages === | |||
| ] | |||
| Common Y-DNA haplogroups in Turkic peoples are ] (found with especially high frequency among Turkic peoples living in present-day ]), ] (especially in ] and, in particular, ]), ] (especially in ] and among ] and the Qangly ]), and ] (especially among Turkic peoples living in present-day ] and the Naiman ]). Some groups also have ] (notably frequent among the ] and ] of Southern Siberia, the ] of the Southern ] region of ], and the ] ]), ] (notably frequent among the ], ], and several other Turkic peoples living in present-day Russia), ] (especially frequent among ], ], and ]), and ] (especially among ], but also observed regularly with low frequency among ], ], ], and ]).<ref>{{Cite journal |vauthors=Zerjal T, Wells RS, Yuldasheva N, Ruzibakiev R, Tyler-Smith C |date=September 2002 |title=A genetic landscape reshaped by recent events: Y-chromosomal insights into central Asia |journal=American Journal of Human Genetics |language=en |volume=71 |issue=3 |pages=466–82 |doi=10.1086/342096 |pmc=419996 |pmid=12145751}}</ref><ref name="Kumar2012">{{Cite book |title=Genomics and Health in the Developing World |vauthors=Kumar D |date=11 May 2012 |publisher=Oxford University Press |isbn=978-0-19-970547-4 |location=Oxford, England |pages=1265–1267}}</ref> | |||
| == Relationship to other Asia-Pacific and Native American populations == | == Relationship to other Asia-Pacific and Native American populations == | ||
| Line 271: | Line 202: | ||
| {{See also|History of Central Asia#Medieval}} | {{See also|History of Central Asia#Medieval}} | ||
| ] | ] | ||
| The genetic evidence suggests that the ] of Central Asia was carried out by ] ] migrating out of Mongolia.<ref>{{Cite journal |display-authors=6 |vauthors=Damgaard PB, Marchi N, Rasmussen S, Peyrot M, Renaud G, Korneliussen T, Moreno-Mayar JV, Pedersen MW, Goldberg A, Usmanova E, Baimukhanov N, Loman V, Hedeager L, Pedersen AG, Nielsen K, Afanasiev G, Akmatov K, Aldashev A, Alpaslan A, Baimbetov G, Bazaliiskii VI, Beisenov A, Boldbaatar B, Boldgiv B, Dorzhu C, Ellingvag S, Erdenebaatar D, Dajani R, Dmitriev E, Evdokimov V, Frei KM, Gromov A, Goryachev A, Hakonarson H, Hegay T, Khachatryan Z, Khaskhanov R, Kitov E, Kolbina A, Kubatbek T, Kukushkin A, Kukushkin I, Lau N, Margaryan A, Merkyte I, Mertz IV, Mertz VK, Mijiddorj E, Moiyesev V, Mukhtarova G, Nurmukhanbetov B, Orozbekova Z, Panyushkina I, Pieta K, Smrčka V, Shevnina I, Logvin A, Sjögren KG, Štolcová T, Taravella AM, Tashbaeva K, Tkachev A, Tulegenov T, Voyakin D, Yepiskoposyan L, Undrakhbold S, Varfolomeev V, Weber A, Wilson Sayres MA, Kradin N, Allentoft ME, Orlando L, Nielsen R, Sikora M, Heyer E, Kristiansen K, Willerslev E |date=May 2018 |title=137 ancient human genomes from across the Eurasian steppes |url=https://www.nature.com/articles/s41586-018-0094-2 |journal=Nature |volume=557 |issue=7705 |pages=369–374 |bibcode=2018Natur.557..369D |doi=10.1038/s41586-018-0094-2 |pmid=29743675 |hdl=1887/3202709 |s2cid=13670282|hdl-access=free |
The genetic evidence suggests that the ] of Central Asia was carried out by ] ] migrating out of Mongolia.<ref>{{Cite journal |display-authors=6 |vauthors=Damgaard PB, Marchi N, Rasmussen S, Peyrot M, Renaud G, Korneliussen T, Moreno-Mayar JV, Pedersen MW, Goldberg A, Usmanova E, Baimukhanov N, Loman V, Hedeager L, Pedersen AG, Nielsen K, Afanasiev G, Akmatov K, Aldashev A, Alpaslan A, Baimbetov G, Bazaliiskii VI, Beisenov A, Boldbaatar B, Boldgiv B, Dorzhu C, Ellingvag S, Erdenebaatar D, Dajani R, Dmitriev E, Evdokimov V, Frei KM, Gromov A, Goryachev A, Hakonarson H, Hegay T, Khachatryan Z, Khaskhanov R, Kitov E, Kolbina A, Kubatbek T, Kukushkin A, Kukushkin I, Lau N, Margaryan A, Merkyte I, Mertz IV, Mertz VK, Mijiddorj E, Moiyesev V, Mukhtarova G, Nurmukhanbetov B, Orozbekova Z, Panyushkina I, Pieta K, Smrčka V, Shevnina I, Logvin A, Sjögren KG, Štolcová T, Taravella AM, Tashbaeva K, Tkachev A, Tulegenov T, Voyakin D, Yepiskoposyan L, Undrakhbold S, Varfolomeev V, Weber A, Wilson Sayres MA, Kradin N, Allentoft ME, Orlando L, Nielsen R, Sikora M, Heyer E, Kristiansen K, Willerslev E |date=May 2018 |title=137 ancient human genomes from across the Eurasian steppes |url=https://www.nature.com/articles/s41586-018-0094-2 |journal=Nature |volume=557 |issue=7705 |pages=369–374 |bibcode=2018Natur.557..369D |doi=10.1038/s41586-018-0094-2 |pmid=29743675 |hdl=1887/3202709 |s2cid=13670282|hdl-access=free}}</ref> According to a recent study, the Turkic Central Asian populations, such as Kyrgyz, Kazakhs, Uzbeks, and Turkmens share more of their gene pool with various East Asian and Siberian populations than with West Asian or European populations. The study further suggests that both migration and linguistic assimilation helped to spread the Turkic languages in Eurasia.<ref>{{Cite journal |display-authors=6 |vauthors=Yunusbayev B, Metspalu M, Metspalu E, Valeev A, Litvinov S, Valiev R, Akhmetova V, Balanovska E, Balanovsky O, Turdikulova S, Dalimova D, Nymadawa P, Bahmanimehr A, Sahakyan H, Tambets K, Fedorova S, Barashkov N, Khidiyatova I, Mihailov E, Khusainova R, Damba L, Derenko M, Malyarchuk B, Osipova L, Voevoda M, Yepiskoposyan L, Kivisild T, Khusnutdinova E, Villems R |date=April 2015 |title=The genetic legacy of the expansion of Turkic-speaking nomads across Eurasia |journal=PLOS Genetics |volume=11 |issue=4 |pages=e1005068 |doi=10.1371/journal.pgen.1005068 |pmc=4405460 |pmid=25898006 |doi-access=free}}</ref> | ||
| === North Asians and Native Americans === | === North Asians and Native Americans === | ||
| Genetic data suggests that ] was populated during the Terminal Upper-Paleolithic (36 |
Genetic data suggests that ] was populated during the Terminal Upper-Paleolithic (36±1.5ka) period from a distinct Paleolithic population migrating through Central Asia into Northern Siberia. This population is known as ] or Ancient North Siberians, who were of West Eurasian origin. | ||
| Between 30,000 and 25,000 years ago, the ancestors of both Paleo-Siberians and Native Americans originated from admixture between ]/Siberians and an Ancient East Asian lineage.<ref>{{cite book | vauthors = Meltzer DJ |title=First Peoples in a New World: Populating Ice Age America |date=2021 |publisher=Cambridge University Press |isbn=978-1-108-49822-7 |page=170 |url=https://books.google.com/books?id=FdE9EAAAQBAJ&pg=PA170 |language=en}}</ref><ref>{{harvnb|Meltzer|2021|p=170|ps=: "The ancestors of Native Americans derive from descendants of Ancient North Siberians and Ancient North Eurasians on the one hand, (exemplified by the genomes of the Yana and Mal'ta individuals, respectively), and a population of Ancient East Asians (signified by the Tianyuan individual). From those groups emerged the deepest ancestors of Native Americans, who became separated far in far northeastern Asia from their Siberian and Asian ancestors around the time of the LGM. Part of the population stayed in Siberia (the Ancient Paleo-Siberians, seen in the Kolyma genome), while others moved east across Beringia."}}</ref> Ancestral Native Americans (or Ancient Beringians) later migrated towards the Beringian region, became isolated from other populations, and subsequently populated the Americas. Further geneflow from ] resulted in the modern distribution of "Neo-Siberians" (associated with |
Between 30,000 and 25,000 years ago, the ancestors of both Paleo-Siberians and Native Americans originated from admixture between ]/Siberians and an Ancient East Asian lineage.<ref>{{cite book | vauthors = Meltzer DJ |title=First Peoples in a New World: Populating Ice Age America |date=2021 |publisher=Cambridge University Press |isbn=978-1-108-49822-7 |page=170 |url=https://books.google.com/books?id=FdE9EAAAQBAJ&pg=PA170 |language=en}}</ref><ref>{{harvnb|Meltzer|2021|p=170|ps=: "The ancestors of Native Americans derive from descendants of Ancient North Siberians and Ancient North Eurasians on the one hand, (exemplified by the genomes of the Yana and Mal'ta individuals, respectively), and a population of Ancient East Asians (signified by the Tianyuan individual). From those groups emerged the deepest ancestors of Native Americans, who became separated far in far northeastern Asia from their Siberian and Asian ancestors around the time of the LGM. Part of the population stayed in Siberia (the Ancient Paleo-Siberians, seen in the Kolyma genome), while others moved east across Beringia."}}</ref> Ancestral Native Americans (or Ancient Beringians) later migrated towards the Beringian region, became isolated from other populations, and subsequently populated the Americas. Further geneflow from ] resulted in the modern distribution of "Neo-Siberians" (associated with ], ], and ] speakers) through the merger of Paleo-Siberians with Northeast Asians.{{sfn|Lazaridis|Belfer-Cohen|Mallick|Patterson|2018}}{{sfn|Moreno-Mayar|Potter|Vinner|Steinrücken|2018}}<ref>{{cite journal | vauthors = Yang MA, Gao X, Theunert C, Tong H, Aximu-Petri A, Nickel B, Slatkin M, Meyer M, Pääbo S, Kelso J, Fu Q | display-authors = 6 | title = 40,000-Year-Old Individual from Asia Provides Insight into Early Population Structure in Eurasia | journal = Current Biology | volume = 27 | issue = 20 | pages = 3202–3208.e9 | date = October 2017 | pmid = 29033327 | pmc = 6592271 | doi = 10.1016/j.cub.2017.09.030 | bibcode = 2017CBio...27E3202Y}}</ref> | ||
| A study found that the North Asian ethnic groups—Altai ], ], ] (]), ], ], ], and ]—are, on average, with the exception of the Komi, more closely related to East Asians than to Europeans, but still occupy a distinct position from the major East Asian populations (typified by Koreans, Japanese, and Han Chinese), who formed a very tight cluster. "Analyses of all 122 populations confirm many known relationships and show that most populations from North Asia form a cluster distinct from all other groups. Refinement of analyses on smaller subsets of populations reinforces the distinctiveness of North Asia and shows that the North Asia cluster identifies a region that is ancestral to Native Americans."<ref>{{cite journal | vauthors = Kidd KK, Evsanaa B, Togtokh A, Brissenden JE, Roscoe JM, Dogan M, Neophytou PI, Gurkan C, Bulbul O, Cherni L, Speed WC, Murtha M, Kidd JR, Pakstis AJ | display-authors = 6 | title = North Asian population relationships in a global context | journal = Scientific Reports | volume = 12 | issue = 1 | pages = 7214 | date = May 2022 | pmid = 35508562 | pmc = 9068624 | doi = 10.1038/s41598-022-10706-x | bibcode = 2022NatSR..12.7214K}}</ref> | |||
| ==== Native Americans ==== | ==== Native Americans ==== | ||
| Multiple studies suggests that all Native Americans ultimately descended from a single founding population that initially diverged from" Ancestral Beringians" which shared a common origin with Paleo-Siberians from the merger of Ancient North Eurasians and a Basal-East Asian source population in Mainland Southeast Asia around 36,000 years ago, at the same time at which the proper ] split from Basal-East Asians, either together or during a separate expansion wave. The basal northern and southern Native American branches, to which all other Indigenous peoples belong, diverged around 16,000 years ago, although earlier dates were also proposed.<ref name="auto3"/><ref name="Moreno-Mayar2018" |
Multiple studies suggests that all Native Americans ultimately descended from a single founding population that initially diverged from" Ancestral Beringians" which shared a common origin with Paleo-Siberians from the merger of Ancient North Eurasians and a Basal-East Asian source population in Mainland Southeast Asia around 36,000 years ago, at the same time at which the proper ] split from Basal-East Asians, either together or during a separate expansion wave. The basal northern and southern Native American branches, to which all other Indigenous peoples belong, diverged around 16,000 years ago, although earlier dates were also proposed.<ref name="auto3" /><ref name="Moreno-Mayar2018" /> An indigenous American sample from 16,000 BCE in ], which is craniometrically similar to modern Native Americans, was found to have been closely related to ], confirming that Ancestral Native Americans split from an ancient Siberian source population somewhere in northeastern Siberia. Genetic data on samples with alleged "Paleo-Indian" morphology turned out to be closely related to contemporary Native Americans, disproving a hypothetical earlier migration into the Americas. The scientists suggest that variation within Native American morphology is just that, the natural variation which have arisen during the formation of Ancestral Native Americans. Signals of a hypothetical "population Y", if not a false positive, are likely explained through a now extinct population from East Asia (e.g. ], which contributed low amounts of ancestry to the Ancestral Native American gene pool in Asia, and perhaps also towards other Asian and Oceanian populations.<ref>{{Cite journal |display-authors=6 |vauthors=Davis LG, Madsen DB, Becerra-Valdivia L, Higham T, Sisson DA, Skinner SM, Stueber D, Nyers AJ, Keen-Zebert A, Neudorf C, Cheyney M, Izuho M, Iizuka F, Burns SR, Epps CW, Willis SC, Buvit I |date=August 2019 |title=Late Upper Paleolithic occupation at Cooper's Ferry, Idaho, USA, ~16,000 years ago |journal=Science |volume=365 |issue=6456 |pages=891–897 |bibcode=2019Sci...365..891D |doi=10.1126/science.aax9830 |pmid=31467216 |doi-access=free}}</ref><ref name="Moreno-Mayar2018" /><ref>{{Cite journal |display-authors=6 |vauthors=Posth C, Nakatsuka N, Lazaridis I, Skoglund P, Mallick S, Lamnidis TC, Rohland N, Nägele K, Adamski N, Bertolini E, Broomandkhoshbacht N, Cooper A, Culleton BJ, Ferraz T, Ferry M, Furtwängler A, Haak W, Harkins K, Harper TK, Hünemeier T, Lawson AM, Llamas B, Michel M, Nelson E, Oppenheimer J, Patterson N, Schiffels S, Sedig J, Stewardson K, Talamo S, Wang CC, Hublin JJ, Hubbe M, Harvati K, Nuevo Delaunay A, Beier J, Francken M, Kaulicke P, Reyes-Centeno H, Rademaker K, Trask WR, Robinson M, Gutierrez SM, Prufer KM, Salazar-García DC, Chim EN, Müller Plumm Gomes L, Alves ML, Liryo A, Inglez M, Oliveira RE, Bernardo DV, Barioni A, Wesolowski V, Scheifler NA, Rivera MA, Plens CR, Messineo PG, Figuti L, Corach D, Scabuzzo C, Eggers S, DeBlasis P, Reindel M, Méndez C, Politis G, Tomasto-Cagigao E, Kennett DJ, Strauss A, Fehren-Schmitz L, Krause J, Reich D |date=November 2018 |title=Reconstructing the Deep Population History of Central and South America |journal=Cell |volume=175 |issue=5 |pages=1185–1197.e22 |doi=10.1016/j.cell.2018.10.027 |pmc=6327247 |pmid=30415837}}</ref><ref>{{Cite journal |vauthors=Willerslev E, Meltzer DJ |date=June 2021 |title=Peopling of the Americas as inferred from ancient genomics |journal=Nature |volume=594 |issue=7863 |pages=356–364 |bibcode=2021Natur.594..356W |doi=10.1038/s41586-021-03499-y |pmid=34135521 |s2cid=235460793}}</ref><ref>{{Cite web | vauthors = Sarkar AA |date=18 June 2021 |title=Ancient Human Genomes Reveal Peopling of the Americas |url=https://www.genengnews.com/news/ancient-human-genomes-reveal-peopling-of-the-americas/ |access-date=15 September 2021 |website=GEN – Genetic Engineering and Biotechnology News |language=en-US |quote=The team discovered that the Spirit Cave remains came from a Native American while dismissing a longstanding theory that a group called Paleoamericans existed in North America before Native Americans.}}</ref> | ||
| === South Asians === | === South Asians === | ||
| {{Main|Genetic history of South Asia}} | {{Main|Genetic history of South Asia}} | ||
| The genetic makeup of modern South Asians can be described as a combination of West Eurasian ancestries with divergent ]. The latter primarily include an indigenous South Asian component (termed ''Ancient Ancestral South Indians'', short "AASI") that is distantly related to the ]s, as well as to ] and ], and further include additional, regionally variable East/Southeast Asian components.<ref>{{Cite journal |display-authors=6 |vauthors=Yelmen B, Mondal M, Marnetto D, Pathak AK, Montinaro F, Gallego Romero I, Kivisild T, Metspalu M, Pagani L |date=August 2019 |title=Ancestry-Specific Analyses Reveal Differential Demographic Histories and Opposite Selective Pressures in Modern South Asian Populations |journal=Molecular Biology and Evolution |volume=36 |issue=8 |pages=1628–1642 |doi=10.1093/molbev/msz037 |pmc=6657728 |pmid=30952160}}</ref><ref name="Yang32" /> The East Asian-related ancestry component forms the major ancestry among ] and ] speakers in the ] and ],<ref name="ChaubeyEast" /><ref name="Chaubey_2010" /> and is generally distributed throughout South Asia at lower frequency, with substantial presence in ].<ref name="ChaubeyEast" /><ref name="Chaubey_2010" /> | |||
| According to a genetic research (2015) including linguistic analyses, suggests an ]n origin for proto-Austroasiatic groups, which first migrated to Southeast Asia and later into India.<ref name="Zhang_2015" |
According to a genetic research (2015) including linguistic analyses, suggests an ]n origin for proto-Austroasiatic groups, which first migrated to Southeast Asia and later into India.<ref name="Zhang_2015" /> According to Ness, there are three broad theories on the origins of the Austroasiatic speakers, namely northeastern India, central or southern China, or southeast Asia.<ref name="Ness-_2014" /> Multiple researches indicate that the Austroasiatic populations in India are derived from (mostly male dominated) migrations from Southeast Asia during the Holocene.<ref name="Chaubey_2011" /><ref name="Zhang_2015" /><ref name="Van_Driem_2007" /><ref name="Riccio2011" /><ref name="ArunKumar_2015" /><!--** START OF NOTE **-->{{refn|"ASI-AAA"|Nevertheless, according to Basu et al. (2016), the AAA were early settlers in India, related to the ASI: "The absence of significant resemblance with any of the neighboring populations is indicative of the ASI and the AAA being early settlers in India, possibly arriving on the "southern exit" wave out of Africa. Differentiation between the ASI and the AAA possibly took place after their arrival in India (ADMIXTURE analysis with K <nowiki>=</nowiki> 3 shows ASI plus AAA to be a single population in SI Appendix, Fig. S2).<ref>{{Cite journal |vauthors=Basu A, Sarkar-Roy N, Majumder PP |date=February 2016 |title=Genomic reconstruction of the history of extant populations of India reveals five distinct ancestral components and a complex structure |journal=Proceedings of the National Academy of Sciences of the United States of America |volume=113 |issue=6 |pages=1594–1599 |bibcode=2016PNAS..113.1594B |doi=10.1073/pnas.1513197113 |pmc=4760789 |pmid=26811443 |doi-access=free}}</ref>{{rp|1598}}}}<!--** END OF NOTE **--> According to Van Driem (2007), "...the mitochondrial picture indicates that the Munda maternal lineage derives from the earliest human settlers on the Subcontinent, whilst the predominant Y chromosome haplogroup argues for a Southeast Asian paternal homeland for Austroasiatic language communities in India."<ref name="Van_Driem_2007" />{{rp|7}} | ||
| According to Chaubey et al. (2011), "Austroasiatic speakers in India today are derived from dispersal from Southeast Asia, followed by extensive sex-specific admixture with local Indian populations."<ref name="Chaubey_2011"/> According to Zhang et al. (2015), Austroasiatic (male) migrations from southeast Asia into India took place after the lates Glacial maximum, circa 4,000 years ago.<ref name="Zhang_2015"/> According to Arunkumar et al. (2015), Y-chromosomal haplogroup O2a1-M95, which is typical for Austroasiatic speaking peoples, clearly decreases from Laos to east India, with "a serial decrease in expansion time from east to west," namely "5.7 ± 0.3 Kya in Laos, 5.2 ± 0.6 in Northeast India, and 4.3 ± 0.2 in East India." This suggests "a late Neolithic east to west spread of the lineage O2a1-M95 from Laos."<ref name="ArunKumar_2015"/><ref>{{Cite magazine | vauthors = Valar M |date=21 April 2015 |title=DNA Reveals Unknown Ancient Migration Into India |url=http://voices.nationalgeographic.com/2015/04/21/genographic-southeast-asia/ |magazine=National Geographic |archive-url=https://web.archive.org/web/20150611183849/http://voices.nationalgeographic.com/2015/04/21/genographic-southeast-asia/ |archive-date=11 June 2015}}</ref> According to Riccio et al. (2011), the ] are likely descended from Austroasiatic migrants from southeast Asia.<ref name="Riccio2011" |
According to Chaubey et al. (2011), "Austroasiatic speakers in India today are derived from dispersal from Southeast Asia, followed by extensive sex-specific admixture with local Indian populations."<ref name="Chaubey_2011" /> According to Zhang et al. (2015), Austroasiatic (male) migrations from southeast Asia into India took place after the lates Glacial maximum, circa 4,000 years ago.<ref name="Zhang_2015" /> According to Arunkumar et al. (2015), Y-chromosomal haplogroup O2a1-M95, which is typical for Austroasiatic speaking peoples, clearly decreases from Laos to east India, with "a serial decrease in expansion time from east to west," namely "5.7 ± 0.3 Kya in Laos, 5.2 ± 0.6 in Northeast India, and 4.3 ± 0.2 in East India." This suggests "a late Neolithic east to west spread of the lineage O2a1-M95 from Laos."<ref name="ArunKumar_2015" /><ref>{{Cite magazine | vauthors = Valar M |date=21 April 2015 |title=DNA Reveals Unknown Ancient Migration Into India |url=http://voices.nationalgeographic.com/2015/04/21/genographic-southeast-asia/ |magazine=National Geographic |archive-url=https://web.archive.org/web/20150611183849/http://voices.nationalgeographic.com/2015/04/21/genographic-southeast-asia/ |archive-date=11 June 2015}}</ref> According to Riccio et al. (2011), the ] are likely descended from Austroasiatic migrants from southeast Asia.<ref name="Riccio2011" /><ref>{{Cite web |title=Austroasiatic Languages |url=http://www.languagesgulper.com/eng/Austroasiatic.html |website=The Language Gulper |vauthors=Gutman A, Avanzati B}}</ref> According to Ness, the Khasi probably migrated into India in the first millennium BCE.<ref name="Ness-_2014" /> | ||
| According to Yelmen et al. 2019, the two main components of Indian genetic variation; the South Asian populations that " |
According to Yelmen et al. 2019, the two main components of Indian genetic variation; the South Asian populations that "separated from East Asian and Andamanese populations" form one of the deepest splits among non-African groups compared to the West Eurasian component because of "40,000 years of independent evolution".<ref>{{cite journal | vauthors = Yelmen B, Mondal M, Marnetto D, Pathak AK, Montinaro F, Gallego Romero I, Kivisild T, Metspalu M, Pagani L | display-authors = 6 | title = Ancestry-Specific Analyses Reveal Differential Demographic Histories and Opposite Selective Pressures in Modern South Asian Populations | journal = Molecular Biology and Evolution | volume = 36 | issue = 8 | pages = 1628–1642 | date = August 2019 | pmid = 30952160 | pmc = 6657728 | doi = 10.1093/molbev/msz037}}</ref> | ||
| Geneflow from Southeast Asians (particularly Austroasiatic groups) to South Asian peoples is associated with the introduction of rice-agriculture to South Asia. There is significant cultural, linguistic, and political Austroasiatic influence on early India, which can also be observed by the presence of Austroasiatic loanwords within ].<ref |
Geneflow from Southeast Asians (particularly Austroasiatic groups) to South Asian peoples is associated with the introduction of rice-agriculture to South Asia. There is significant cultural, linguistic, and political Austroasiatic influence on early India, which can also be observed by the presence of Austroasiatic loanwords within ].<ref>{{Cite book | vauthors = Lévi S, Przyluski J, Bloch J |url=https://books.google.com/books?id=dx5dzJGGBg0C&q=austroasiatic+influence+on+india&pg=PR15 |title=Pre-Aryan and Pre-Dravidian in India |date=1993 |publisher=Asian Educational Services |isbn=978-81-206-0772-9 |language=en |quote=It has been further proved that not only linguistic but also certain cultural and political facts of ancient India, can be explained by Austroasiatic elements.}}</ref><ref>{{Cite web |title=How rice farming may have spread across the ancient world |url=https://www.science.org/content/article/how-rice-farming-may-have-spread-across-ancient-world |access-date=26 October 2021 |website=science.org |language=en}}</ref> | ||
| === Southeast Asians === | === Southeast Asians === | ||
| ] | ] | ||
| A 2020 genetic study about Southeast Asian populations, found that mostly all Southeast Asians are closely related to East Asians and have mostly "East Asian-related" ancestry.<ref name="auto5"/><ref>{{cite journal | vauthors = Liu D, Duong NT, Ton ND, Van Phong N, Pakendorf B, Van Hai N, Stoneking M | title = Extensive Ethnolinguistic Diversity in Vietnam Reflects Multiple Sources of Genetic Diversity | journal = Molecular Biology and Evolution | volume = 37 | issue = 9 | pages = 2503–2519 | date = September 2020 | pmid = 32344428 | pmc = 7475039 | doi = 10.1093/molbev/msaa099 | doi-access = free |
A 2020 genetic study about Southeast Asian populations, found that mostly all Southeast Asians are closely related to East Asians and have mostly "East Asian-related" ancestry.<ref name="auto5" /><ref>{{cite journal | vauthors = Liu D, Duong NT, Ton ND, Van Phong N, Pakendorf B, Van Hai N, Stoneking M | title = Extensive Ethnolinguistic Diversity in Vietnam Reflects Multiple Sources of Genetic Diversity | journal = Molecular Biology and Evolution | volume = 37 | issue = 9 | pages = 2503–2519 | date = September 2020 | pmid = 32344428 | pmc = 7475039 | doi = 10.1093/molbev/msaa099 | doi-access = free}}</ref> | ||
| Ancient remains of hunter-gatherers in Maritime Southeast Asia, such as one Holocene hunter-gatherer from ], had ancestry from both, |
Ancient remains of hunter-gatherers in Maritime Southeast Asia, such as one Holocene hunter-gatherer from ], had ancestry from both, an Australasian lineage (represented by Papuans and Aboriginal Australasians) and an "Ancient Asian" lineage (represented by East Asians or Andamanese Onge). The hunter-gatherer individual had approximately c. 50% "Basal-East Asian" ancestry and c. 50% Australasian/Papuan ancestry, and was positioned in between modern East Asians and Papuans of Oceania. The authors concluded that East Asian-related ancestry expanded from ] into ] much earlier than previously suggested, as early as 25,000 BCE, long before the expansion of ] and ] groups.<ref>{{Cite journal |display-authors=6 |vauthors=Carlhoff S, Duli A, Nägele K, Nur M, Skov L, Sumantri I, Oktaviana AA, Hakim B, Burhan B, Syahdar FA, McGahan DP, Bulbeck D, Perston YL, Newman K, Saiful AM, Ririmasse M, Chia S, Pulubuhu DA, Jeong C, Peter BM, Prüfer K, Powell A, Krause J, Posth C, Brumm A |date=August 2021 |title=Genome of a middle Holocene hunter-gatherer from Wallacea |journal=Nature |volume=596 |issue=7873 |pages=543–547 |bibcode=2021Natur.596..543C |doi=10.1038/s41586-021-03823-6 |pmc=8387238 |pmid=34433944 |quote=The qpGraph analysis confirmed this branching pattern, with the Leang Panninge individual branching off from the Near Oceanian clade after the Denisovan gene flow, although with the most supported topology indicating around 50% of a basal East Asian component contributing to the Leang Panninge genome (Fig. 3c, Supplementary Figs. 7–11). |hdl-access=free |hdl=10072/407535}}</ref> | ||
| A 2022 genetic study confirmed the close link between East Asians and Southeast Asians, which the authors term "East/Southeast Asian" (ESEA) populations, and also found a low but consistent proportion of South Asian-associated "SAS ancestry" (best samplified by modern ] from ]) among specific Mainland Southeast Asian (MESA) ethnic groups (~2–16% as inferred by ''qpAdm''), likely as a result of cultural diffision; mainly of South Asian merchants spreading Hinduism and Buddhism among the Indianized kingdoms of Southeast Asia. The authors however caution that Bengali samples harbor |
A 2022 genetic study confirmed the close link between East Asians and Southeast Asians, which the authors term "East/Southeast Asian" (ESEA) populations, and also found a low but consistent proportion of South Asian-associated "SAS ancestry" (best samplified by modern ] from ]) among specific Mainland Southeast Asian (MESA) ethnic groups (~2–16% as inferred by ''qpAdm''), likely as a result of cultural diffision; mainly of South Asian merchants spreading Hinduism and Buddhism among the Indianized kingdoms of Southeast Asia. The authors however caution that Bengali samples harbor detectable East Asian ancestry, which may affect the estimation of shared haplotypes. Overall, the geneflow event is estimated to have happened between 500 and 1000 YBP.<ref>{{cite journal | vauthors = Changmai P, Jaisamut K, Kampuansai J, Kutanan W, Altınışık NE, Flegontova O, Inta A, Yüncü E, Boonthai W, Pamjav H, Reich D, Flegontov P | display-authors = 6 | title = Indian genetic heritage in Southeast Asian populations | journal = PLOS Genetics | volume = 18 | issue = 2 | pages = e1010036 | date = February 2022 | pmid = 35176016 | pmc = 8853555 | doi = 10.1371/journal.pgen.1010036 | doi-access = free}}</ref> | ||
| ] | |||
| === |
=== Australasians === | ||
| {{Main|Indigenous people of New Guinea|Melanesians|Aboriginal Australians}} | |||
| Genetic studies have revealed that Aboriginal Australians largely descended from an Eastern Eurasian population wave, and are most closely related to other ]ns, such as ] such as ] and also show affinity to other Asian and Pacific populations, such as ], Ancient Ancestral South Indians and East Asian peoples. Phylogenetic data suggests that an early initial eastern non-African lineage (ENA) or East-Eurasian meta-population trifurcated, and gave rise to Australasians (Oceanians), the Ancient Ancestral South Indians/Andamanese, and the East/Southeast Asian lineage including the ancestors of ].<ref name="Yang"/> | |||
| ] and ] are deeply related to East Asians. Genetic studies have revealed that Australasians descended from the same Eastern Eurasian source population as East Asians and indigenous South Asians (AASI).<ref name="Yang32" /> | |||
| {{clear}} | |||
| === Melanesians === | |||
| Phylogenetic data suggests that an early initial Eastern Eurasian (eastern non-African ENA) meta-population trifurcated, and gave rise to Australasians (Oceanians), the AASI, as well as East/Southeast Asians, although Papuans may have also received some geneflow from an earlier group (xOoA),<ref name="auto6"/><ref name="Yang"/> around 2%,<ref name="auto7"/> next to additional archaic admixture in the ] region. Vallini et al. 2022 study confirmed the position of Papuans, who belong to the broader ] peoples of the Pacific Islands, as a sister lineage of East Asians: Papuan ancestry can be modelled either as an almost unadmixed sister lineage of East Asians (with potential minimal ingressions from basal Out-of-Africa populations) or as an approximately even mixture of East Asians and a basal East-Eurasian lineage that had split prior to the expansion of East-Eurasian ancestry into East Asia and Oceania.<ref name="auto6" /> | |||
| == Notes == | |||
| {{reflist|group=note}} | |||
| == References == | == References == | ||
| {{reflist|refs= | {{reflist|refs= | ||
| <ref name="gene">{{cite journal |doi=10.1038/nature02878 |title=Genetic evidence supports demic diffusion of Han culture |year=2004 |last1=Wen |first1=Bo |last2=Li |first2=Hui |last3=Lu |first3=Daru |last4=Song |first4=Xiufeng |last5=Zhang |first5=Feng |last6=He |first6=Yungang |last7=Li |first7=Feng |last8=Gao |first8=Yang |last9=Mao |first9=Xianyun|last10=Zhang |first10=L |last11=Qian |first11=J |last12=Tan |first12=J |last13=Jin |first13=J |last14=Huang |first14=W |last15=Deka |first15=R |last16=Su |first16=B |last17=Chakraborty |first17=R |last18=Jin |first18=L |display-authors=9|journal=Nature |volume=431 |issue=7006 |pages=302–05 |pmid=15372031|bibcode=2004Natur.431..302W |s2cid=4301581}}</ref> | |||
| <ref name=":6">{{cite journal | vauthors = Yang MA, Fan X, Sun B, Chen C, Lang J, Ko YC, Tsang CH, Chiu H, Wang T, Bao Q, Wu X, Hajdinjak M, Ko AM, Ding M, Cao P, Yang R, Liu F, Nickel B, Dai Q, Feng X, Zhang L, Sun C, Ning C, Zeng W, Zhao Y, Zhang M, Gao X, Cui Y, Reich D, Stoneking M, Fu Q | display-authors = 6 | title = Ancient DNA indicates human population shifts and admixture in northern and southern China | journal = Science | volume = 369 | issue = 6501 | pages = 282–288 | date = July 2020 | pmid = 32409524 | doi = 10.1126/science.aba0909 | s2cid = 218649510 | bibcode = 2020Sci...369..282Y}}</ref> | |||
| {{cite journal |display-authors=6 |vauthors=Srithawong S, Srikummool M, Pittayaporn P, Ghirotto S, Chantawannakul P, Sun J, Eisenberg A, Chakraborty R, Kutanan W |date=July 2015 |title=Genetic and linguistic correlation of the Kra-Dai-speaking groups in Thailand |journal=Journal of Human Genetics |volume=60 |issue=7 |pages=371–80 |doi=10.1038/jhg.2015.32 |pmid=25833471 |s2cid=21509343 |doi-access=free}} | |||
| <ref name=":6">{{cite journal | vauthors = Yang MA, Fan X, Sun B, Chen C, Lang J, Ko YC, Tsang CH, Chiu H, Wang T, Bao Q, Wu X, Hajdinjak M, Ko AM, Ding M, Cao P, Yang R, Liu F, Nickel B, Dai Q, Feng X, Zhang L, Sun C, Ning C, Zeng W, Zhao Y, Zhang M, Gao X, Cui Y, Reich D, Stoneking M, Fu Q | display-authors = 6 | title = Ancient DNA indicates human population shifts and admixture in northern and southern China | journal = Science | volume = 369 | issue = 6501 | pages = 282–288 | date = July 2020 | pmid = 32409524 | doi = 10.1126/science.aba0909 | s2cid = 218649510 | bibcode = 2020Sci...369..282Y }}</ref> | |||
| <ref name="Ancient Jomon genome sequence analy">{{cite journal |display-authors=6 |vauthors=Gakuhari T, Nakagome S, Rasmussen S, Allentoft ME, Sato T, Korneliussen T, Chuinneagáin BN, Matsumae H, Koganebuchi K, Schmidt R, Mizushima S, Kondo O, Shigehara N, Yoneda M, Kimura R, Ishida H, Masuyama T, Yamada Y, Tajima A, Shibata H, Toyoda A, Tsurumoto T, Wakebe T, Shitara H, Hanihara T, Willerslev E, Sikora M, Oota H |date=August 2020 |title=Ancient Jomon genome sequence analysis sheds light on migration patterns of early East Asian populations |journal=Communications Biology |volume=3 |issue=1 |pages=437 |doi=10.1038/s42003-020-01162-2 |pmc=7447786 |pmid=32843717 |quote=Population genomic studies on present-day humans7,8 have exclusively supported the southern route origin of East Asian populations.}}</ref> | <ref name="Ancient Jomon genome sequence analy">{{cite journal |display-authors=6 |vauthors=Gakuhari T, Nakagome S, Rasmussen S, Allentoft ME, Sato T, Korneliussen T, Chuinneagáin BN, Matsumae H, Koganebuchi K, Schmidt R, Mizushima S, Kondo O, Shigehara N, Yoneda M, Kimura R, Ishida H, Masuyama T, Yamada Y, Tajima A, Shibata H, Toyoda A, Tsurumoto T, Wakebe T, Shitara H, Hanihara T, Willerslev E, Sikora M, Oota H |date=August 2020 |title=Ancient Jomon genome sequence analysis sheds light on migration patterns of early East Asian populations |journal=Communications Biology |volume=3 |issue=1 |pages=437 |doi=10.1038/s42003-020-01162-2 |pmc=7447786 |pmid=32843717 |quote=Population genomic studies on present-day humans7,8 have exclusively supported the southern route origin of East Asian populations.}}</ref> | ||
| <ref name="auto1">{{cite journal |display-authors=6 |vauthors=Yang MA, Fan X, Sun B, Chen C, Lang J, Ko YC, Tsang CH, Chiu H, Wang T, Bao Q, Wu X, Hajdinjak M, Ko AM, Ding M, Cao P, Yang R, Liu F, Nickel B, Dai Q, Feng X, Zhang L, Sun C, Ning C, Zeng W, Zhao Y, Zhang M, Gao X, Cui Y, Reich D, Stoneking M, Fu Q |date=July 2020 |title=Ancient DNA indicates human population shifts and admixture in northern and southern China |journal=Science |volume=369 |issue=6501 |pages=282–288 |bibcode=2020Sci...369..282Y |doi=10.1126/science.aba0909 |pmid=32409524 |s2cid=218649510}}</ref> | |||
| <ref name="Niall22">{{cite journal | vauthors = Cooke NP, Mattiangeli V, Cassidy LM, Okazaki K, Stokes CA, Onbe S, Hatakeyama S, Machida K, Kasai K, Tomioka N, Matsumoto A, Ito M, Kojima Y, Bradley DG, Gakuhari T, Nakagome S | display-authors = 6 | title = Ancient genomics reveals tripartite origins of Japanese populations | journal = Science Advances | volume = 7 | issue = 38 | pages = eabh2419 | date = September 2021 | pmid = 34533991 | pmc = 8448447 | doi = 10.1126/sciadv.abh2419 | bibcode = 2021SciA....7.2419C }}</ref> | |||
| <ref name="auto3">{{cite journal |display-authors=6 |vauthors=Gakuhari T, Nakagome S, Rasmussen S, Allentoft ME, Sato T, Korneliussen T, Chuinneagáin BN, Matsumae H, Koganebuchi K, Schmidt R, Mizushima S, Kondo O, Shigehara N, Yoneda M, Kimura R, Ishida H, Masuyama T, Yamada Y, Tajima A, Shibata H, Toyoda A, Tsurumoto T, Wakebe T, Shitara H, Hanihara T, Willerslev E, Sikora M, Oota H |date=August 2020 |title=Ancient Jomon genome sequence analysis sheds light on migration patterns of early East Asian populations |journal=Communications Biology |volume=3 |issue=1 |pages=437 |doi=10.1038/s42003-020-01162-2 |pmc=7447786 |pmid=32843717}}</ref> | <ref name="auto3">{{cite journal |display-authors=6 |vauthors=Gakuhari T, Nakagome S, Rasmussen S, Allentoft ME, Sato T, Korneliussen T, Chuinneagáin BN, Matsumae H, Koganebuchi K, Schmidt R, Mizushima S, Kondo O, Shigehara N, Yoneda M, Kimura R, Ishida H, Masuyama T, Yamada Y, Tajima A, Shibata H, Toyoda A, Tsurumoto T, Wakebe T, Shitara H, Hanihara T, Willerslev E, Sikora M, Oota H |date=August 2020 |title=Ancient Jomon genome sequence analysis sheds light on migration patterns of early East Asian populations |journal=Communications Biology |volume=3 |issue=1 |pages=437 |doi=10.1038/s42003-020-01162-2 |pmc=7447786 |pmid=32843717}}</ref> | ||
| <ref name="auto4">{{cite journal |display-authors=6 |vauthors=Larena M, Sanchez-Quinto F, Sjödin P, McKenna J, Ebeo C, Reyes R, Casel O, Huang JY, Hagada KP, Guilay D, Reyes J, Allian FP, Mori V, Azarcon LS, Manera A, Terando C, Jamero L, Sireg G, Manginsay-Tremedal R, Labos MS, Vilar RD, Latiph A, Saway RL, Marte E, Magbanua P, Morales A, Java I, Reveche R, Barrios B, Burton E, Salon JC, Kels MJ, Albano A, Cruz-Angeles RB, Molanida E, Granehäll L, Vicente M, Edlund H, Loo JH, Trejaut J, Ho SY, Reid L, Malmström H, Schlebusch C, Lambeck K, Endicott P, Jakobsson M |date=March 2021 |title=Multiple migrations to the Philippines during the last 50,000 years |journal=Proceedings of the National Academy of Sciences of the United States of America |volume=118 |issue=13 |pages=e2026132118 |doi=10.1073/pnas.2026132118 |pmc=8020671 |pmid=33753512 |bibcode=2021PNAS..11826132L |doi-access=free}}</ref> | <ref name="auto4">{{cite journal |display-authors=6 |vauthors=Larena M, Sanchez-Quinto F, Sjödin P, McKenna J, Ebeo C, Reyes R, Casel O, Huang JY, Hagada KP, Guilay D, Reyes J, Allian FP, Mori V, Azarcon LS, Manera A, Terando C, Jamero L, Sireg G, Manginsay-Tremedal R, Labos MS, Vilar RD, Latiph A, Saway RL, Marte E, Magbanua P, Morales A, Java I, Reveche R, Barrios B, Burton E, Salon JC, Kels MJ, Albano A, Cruz-Angeles RB, Molanida E, Granehäll L, Vicente M, Edlund H, Loo JH, Trejaut J, Ho SY, Reid L, Malmström H, Schlebusch C, Lambeck K, Endicott P, Jakobsson M |date=March 2021 |title=Multiple migrations to the Philippines during the last 50,000 years |journal=Proceedings of the National Academy of Sciences of the United States of America |volume=118 |issue=13 |pages=e2026132118 |doi=10.1073/pnas.2026132118 |pmc=8020671 |pmid=33753512 |bibcode=2021PNAS..11826132L |doi-access=free}}</ref> | ||
| <ref name="Gan2008">{{cite journal |vauthors=Gan RJ, Pan SL, Mustavich LF, Qin ZD, Cai XY, Qian J, Liu CW, Peng JH, Li SL, Xu JS, Jin L, Li H |year=2008 |title=Pinghua population as an exception of Han Chinese's coherent genetic structure |journal=Journal of Human Genetics |volume=53 |issue=4 |pages=303–13 |doi=10.1007/s10038-008-0250-x |pmid=18270655 |doi-access=free}}</ref> | <ref name="Gan2008">{{cite journal |vauthors=Gan RJ, Pan SL, Mustavich LF, Qin ZD, Cai XY, Qian J, Liu CW, Peng JH, Li SL, Xu JS, Jin L, Li H |year=2008 |title=Pinghua population as an exception of Han Chinese's coherent genetic structure |journal=Journal of Human Genetics |volume=53 |issue=4 |pages=303–13 |doi=10.1007/s10038-008-0250-x |pmid=18270655 |doi-access=free}}</ref> | ||
| <ref name="Jin">{{cite journal | vauthors = Jin HJ, Tyler-Smith C, Kim W | title = The peopling of Korea revealed by analyses of mitochondrial DNA and Y-chromosomal markers | journal = PLOS ONE | volume = 4 | issue = 1 | pages = e4210 | date = 16 January 2009 | pmid = 19148289 | pmc = 2615218 | doi = 10.1371/journal.pone.0004210 | bibcode = 2009PLoSO...4.4210J | doi-access = free}}</ref> | |||
| {{ |
<ref name="Kim2011">{{Cite journal |vauthors=Kim SH, Kim KC, Shin DJ, Jin HJ, Kwak KD, Han MS, Song JM, Kim W, Kim W |date=April 2011 |title=High frequencies of Y-chromosome haplogroup O2b-SRY465 lineages in Korea: a genetic perspective on the peopling of Korea |journal=Investigative Genetics |volume=2 |issue=1 |pages=10 |doi=10.1186/2041-2223-2-10 |pmc=3087676 |pmid=21463511 |doi-access=free}} ] Text was copied from this source, which is available under a license.</ref> | ||
| <ref name="Sato evab192">{{cite journal | vauthors = Sato T, Adachi N, Kimura R, Hosomichi K, Yoneda M, Oota H, Tajima A, Toyoda A, Kanzawa-Kiriyama H, Matsumae H, Koganebuchi K, Shimizu KK, Shinoda KI, Hanihara T, Weber A, Kato H, Ishida H | display-authors = 6 | title = Whole-Genome Sequencing of a 900-Year-Old Human Skeleton Supports Two Past Migration Events from the Russian Far East to Northern Japan | journal = Genome Biology and Evolution | volume = 13 | issue = 9 | pages = evab192 | date = September 2021 | pmid = 34410389 | pmc = 8449830 | doi = 10.1093/gbe/evab192}}</ref> | |||
| <ref name=":2">{{Cite journal |vauthors=Osada N, Kawai Y |date=2021 |title=Exploring models of human migration to the Japanese archipelago using genome-wide genetic data |url=https://www.jstage.jst.go.jp/article/ase/129/1/129_201215/_html/-char/en |journal=Anthropological Science |volume=129 |issue=1 |pages=45–58 |doi=10.1537/ase.201215 |quote=Via the southern route, ancestors of current Asian populations reached Southeast Asia and a part of Oceania around 70000–50000 years ago, probably through a coastal dispersal route (Bae et al., 2017). The oldest samples providing the genetic evidence of the northern migration route come from a high-coverage genome sequence of individuals excavated from the Yana RHS site in northeastern Siberia (Figure 2), which is about 31600 years old (Sikora et al., 2019). A wide range of artifacts, including bone crafts of wooly rhinoceros and mammoths, were excavated at the site (Pitulko et al., 2004). The analysis of genome sequences showed that the samples were deeply diverged from most present-day East Asians and more closely related to present-day Europeans, suggesting that the population reached the area through a route different from the southern route. A 24000-year-old individual excavated near Lake Baikal (Figure 2), also known as the Mal'ta boy, and 17000-year-old individuals from the Afontova Gora II site (Afontova Gora 2 and 3) showed similar genetic features to the Yana individuals (Raghavan et al., 2014; Fu et al., 2016; Sikora et al., 2019). Interestingly, genetic data suggested that Yana individuals received a large amount of gene flow from the East Asian lineage (Sikora et al., 2019; Yang et al., 2020). |s2cid=234247309|doi-access=free}}</ref> | |||
| <ref name="Hammer2006">{{cite journal |vauthors=Hammer MF, Karafet TM, Park H, Omoto K, Harihara S, Stoneking M, Horai S |year=2006 |title=Dual origins of the Japanese: common ground for hunter-gatherer and farmer Y chromosomes |journal=Journal of Human Genetics |volume=51 |issue=1 |pages=47–58 |doi=10.1007/s10038-005-0322-0 |pmid=16328082 |doi-access=free}}</ref> | |||
| <ref name="Yang32">{{Cite journal |vauthors=Yang MA |date=6 January 2022 |title=A genetic history of migration, diversification, and admixture in Asia |url=http://www.pivotscipub.com/hpgg/2/1/0001 |journal=Human Population Genetics and Genomics |language=en |volume=2 |issue=1 |pages=1–32 |doi=10.47248/hpgg2202010001 |issn=2770-5005 |quote=...In contrast, mainland East and Southeast Asians and other Pacific islanders (e.g., Austronesian speakers) are closely related to each other and here denoted as belonging to an East and Southeast Asian (ESEA) lineage (Box 2). …the ESEA lineage differentiated into at least three distinct ancestries: Tianyuan ancestry which can be found 40,000–33,000 years ago in northern East Asia, ancestry found today across present-day populations of East Asia, Southeast Asia, and Siberia, but whose origins are unknown, and Hòabìnhian ancestry found 8,000–4,000 years ago in Southeast Asia, but whose origins in the Upper Paleolithic are unknown. |doi-access=free}}</ref> | |||
| {{cite journal |vauthors= |
<ref name="auto5">{{cite journal |vauthors=Tagore D, Aghakhanian F, Naidu R, Phipps ME, Basu A |date=March 2021 |title=Insights into the demographic history of Asia from common ancestry and admixture in the genomic landscape of present-day Austroasiatic speakers |journal=BMC Biology |volume=19 |issue=1 |pages=61 |doi=10.1186/s12915-021-00981-x |pmc=8008685 |pmid=33781248 |doi-access=free}}</ref> | ||
| <ref name="sciencedirect.com">{{Cite journal |last1=Zhang |first1=Ming |last2=Fu |first2=Qiaomei |date=1 June 2020 |title=Human evolutionary history in Eastern Eurasia using insights from ancient DNA |url=https://www.sciencedirect.com/science/article/pii/S0959437X2030109X |journal=Current Opinion in Genetics & Development |series=Genetics of Human Origin |volume=62 |pages=78–84 |doi=10.1016/j.gde.2020.06.009 |pmid=32688244 |s2cid=220671047 |issn=0959-437X}}</ref> | |||
| <ref name="Savelyev & Jeong, 2020">{{cite journal |last1=Savelyev |first1=Alexander |last2=Jeong |first2=Choongwoon |date=7 May 2020 |title=Early nomads of the Eastern Steppe and their tentative connections in the West |journal=] |volume=2 |issue=E20 |doi=10.1017/ehs.2020.18 |pmc=7612788 |pmid=35663512 |s2cid=218935871 |hdl=21.11116/0000-0007-772B-4}} ] Text was copied from this source, which is available under a ]. "Such a distribution of Xiongnu words may be an indication that both Turkic and Eastern Iranian-speaking groups were present among the Xiongnu in the earlier period of their history. Etymological analysis shows that some crucial components in the Xiongnu political, economic and cultural package, including dairy pastoralism and elements of state organization, may have been imported by the Eastern Iranians. Arguably, these Iranian-speaking groups were assimilated over time by the predominant Turkic-speaking part of the Xiongnu population. ... The genetic profile of published Xiongnu individuals speaks against the Yeniseian hypothesis, assuming that modern Yeniseian speakers (i.e. Kets) are representative of the ancestry components in the historical Yeniseian speaking groups in southern Siberia. In contrast to the Iron Age populations listed in Table 2, Kets do not have the Iranian-related ancestry component but harbour a strong genetic affinity with Samoyedic-speaking neighbours, such as Selkups (Jeong et al., 2018, 2019)."</ref> | |||
| <ref name="Jin">{{cite journal | vauthors = Jin HJ, Tyler-Smith C, Kim W | title = The peopling of Korea revealed by analyses of mitochondrial DNA and Y-chromosomal markers | journal = PLOS ONE | volume = 4 | issue = 1 | pages = e4210 | date = 16 January 2009 | pmid = 19148289 | pmc = 2615218 | doi = 10.1371/journal.pone.0004210 | bibcode = 2009PLoSO...4.4210J | doi-access = free }}</ref> | |||
| <ref name=":92">{{Cite journal |last1=Cooke |first1=Niall P. |last2=Mattiangeli |first2=Valeria |last3=Cassidy |first3=Lara M. |last4=Okazaki |first4=Kenji |last5=Stokes |first5=Caroline A. |last6=Onbe |first6=Shin |last7=Hatakeyama |first7=Satoshi |last8=Machida |first8=Kenichi |last9=Kasai |first9=Kenji |last10=Tomioka |first10=Naoto |last11=Matsumoto |first11=Akihiko |date=September 2021 |title=Ancient genomics reveals tripartite origins of Japanese populations |journal=Science Advances |language=EN |volume=7 |issue=38 |pages=eabh2419 |bibcode=2021SciA....7.2419C |doi=10.1126/sciadv.abh2419 |pmc=8448447 |pmid=34533991}}</ref> | |||
| <ref name="Xue2006">{{cite journal |last1=Xue |first1=Yali |last2=Zerjal |first2=Tatiana |last3=Bao |first3=Weidong |last4=Zhu |first4=Suling |last5=Shu |first5=Qunfang |last6=Xu |first6=Jiujin |last7=Du |first7=Ruofu |last8=Fu |first8=Songbin |last9=Li |first9=Pu |last10=Hurles |first10=Matthew E |last11=Yang |first11=Huanming |last12=Tyler-Smith |first12=Chris |title=Male Demography in East Asia: A North–South Contrast in Human Population Expansion Times |journal=Genetics |date=1 April 2006 |volume=172 |issue=4 |pages=2431–2439 |doi=10.1534/genetics.105.054270 |pmid=16489223 |pmc=1456369}}</ref> | |||
| {{cite journal |url=http://mbe.oxfordjournals.org/content/27/8/1833 |vauthors=Karafet TM, Hallmark B, Cox MP, Sudoyo H, Downey S, Lansing JS, Hammer MF |date=August 2010 |title=Major East–West Division Underlies Y Chromosome Stratification across Indonesia |journal=Molecular Biology and Evolution |volume=27 |issue=8 |pages=1833–1844 |doi=10.1093/molbev/msq063 |pmid=20207712 |doi-access=free}} | |||
| <ref name="Wang2018">Wang Chi-zao, Shi Mei-sen, and Li Hui, "The Origin of Daur from the Perspective of Molecular Anthropology." ''Journal of North Minzu University'' 2018, No.5, Gen.No.143.</ref> | |||
| <ref name="DidYouKnowDNA">{{Cite web |date=14 November 2016 |title=Did you know DNA was used to uncover the origin of the House of Aisin Gioro? |url=https://www.didyouknowdna.com/famous-dna/aisin-gioro-dna/ |access-date=5 November 2020 |website=Did You Know DNA...}}</ref> | |||
| <ref name="Kim2011">{{Cite journal |vauthors=Kim SH, Kim KC, Shin DJ, Jin HJ, Kwak KD, Han MS, Song JM, Kim W, Kim W |date=April 2011 |title=High frequencies of Y-chromosome haplogroup O2b-SRY465 lineages in Korea: a genetic perspective on the peopling of Korea |journal=Investigative Genetics |volume=2 |issue=1 |pages=10 |doi=10.1186/2041-2223-2-10 |pmc=3087676 |pmid=21463511 |doi-access=free }} ] Text was copied from this source, which is available under a license.</ref> | |||
| <ref name="Suzuki2">{{cite journal |last=Suzuki |first=Yuka |date=6 December 2012 |title=Ryukyuan, Ainu People Genetically Similar |url=http://www.asianscientist.com/2012/12/in-the-lab/ryukyuan-ainu-people-genetically-similar-2012/ |url-status=live |journal=Asian Scientist |archive-url=https://web.archive.org/web/20160616171127/http://www.asianscientist.com/2012/12/in-the-lab/ryukyuan-ainu-people-genetically-similar-2012/ |archive-date=16 June 2016 |access-date=11 June 2016}}</ref> | |||
| {{ |
<ref name="Wang 2021 413–419">{{Cite journal |last=Wang |first=Chuan-Chao |date=2021 |title=Genomic insights into the formation of human populations in East Asia |url=https://www.researchgate.net/publication/349510968 |journal=Nature |volume=591 |issue=7850 |pages=413–419 |bibcode=2021Natur.591..413W |doi=10.1038/s41586-021-03336-2 |pmc=7993749 |pmid=33618348}}</ref> | ||
| <ref name=":42">{{cite journal |last1=Wang |first1=Rui |last2=Wang |first2=Chuan-Chao |date=8 August 2022 |title=Human genetics: The dual origin of Three Kingdoms period Koreans |journal=Current Biology |language=en |volume=32 |issue=15 |pages=R844–R847 |doi=10.1016/j.cub.2022.06.044 |issn=0960-9822 |pmid=35944486 |s2cid=251410856 |doi-access=free|bibcode=2022CBio...32.R844W}}</ref> | |||
| <ref name="Kwon2015">So Yeun Kwon, Hwan Young Lee, Eun Young Lee, Woo Ick Yang, and Kyoung-Jin Shin, "Confirmation of Y haplogroup tree topologies with newly suggested Y-SNPs for the C2, O2b and O3a subhaplogroups." ''Forensic Science International: Genetics'' 19 (2015) 42–46. http://dx.doi.org/10.1016/j.fsigen.2015.06.003</ref> | |||
| {{cite journal |vauthors=Ochiai E, Minaguchi K, Nambiar P, Kakimoto Y, Satoh F, Nakatome M, Miyashita K, Osawa M |date=September 2016 |title=Evaluation of Y chromosomal SNP haplogrouping in the HID-Ion AmpliSeq™ Identity Panel |journal=Legal Medicine |volume=22 |pages=58–61 |doi=10.1016/j.legalmed.2016.08.001 |pmid=27591541}} | |||
| <!-- | |||
| <ref name=" |
<ref name="pmid 29636655">{{Cite journal |last1=Wang |first1=Yuchen |last2= Lu |first2=Dongsheng |last3=Chung |first3=Yeun-Jun |last4=Xu |first4=Shuhua |title=Genetic structure, divergence and admixture of Han Chinese, Japanese and Korean populations |journal=Hereditas |volume=155 |page=19 |publication-date=6 April 2018|doi=10.1186/s41065-018-0057-5 |pmid=29636655 |pmc=5889524 |year=2018 |doi-access=free}}</ref> | ||
| --> | |||
| <ref name="He2023">{{cite journal |last1=He |first1=Guanglin |last2=Wang |first2=Mengge |last3=Miao |first3=Lei |last4=Chen |first4=Jing |last5=Zhao |first5=Jie |last6=Sun |first6=Qiuxia |last7=Duan |first7=Shuhan |last8=Wang |first8=Zhiyong |last9=Xu |first9=Xiaofei |last10=Sun |first10=Yuntao |last11=Liu |first11=Yan |last12=Liu |first12=Jing |last13=Wang |first13=Zheng |last14=Wei |first14=Lanhai |last15=Liu |first15=Chao |last16=Ye |first16=Jian |last17=Wang |first17=Le |title=Multiple founding paternal lineages inferred from the newly-developed 639-plex Y-SNP panel suggested the complex admixture and migration history of Chinese people |journal=Human Genomics |date=28 March 2023 |volume=17 |issue=1 |page=29 |doi=10.1186/s40246-023-00476-6 |doi-access=free |pmid=36973821 |pmc=10045532}}</ref> | |||
| {{cite journal |vauthors=Sato Y, Shinka T, Ewis AA, Yamauchi A, Iwamoto T, Nakahori Y |year=2014 |title=Overview of genetic variation in the Y chromosome of modern Japanese males |journal=Anthropological Science |volume=122 |issue=3 |pages=131–136 |doi=10.1537/ase.140709 |issn=0918-7960 |doi-access=free}} | |||
| <ref name="Lu2009">{{cite journal | last1 = Lu | first1 = Chuncheng | last2 = Zhang | first2 = Jie | last3 = Li | first3 = Yingchun | last4 = Xia | first4 = Yankai | last5 = Zhang | first5 = Feng | last6 = Wu | first6 = Bin | last7 = Wu | first7 = Wei | last8 = Ji | first8 = Guixiang | last9 = Gu | first9 = Aihua | last10 = Wang | first10 = Shoulin | last11 = Jin | first11 = Li | last12 = Wang | first12 = Xinru | year = 2009 | title = The b2/b3 subdeletion shows higher risk of spermatogenic failure and higher frequency of complete AZFc deletion than the gr/gr subdeletion in a Chinese population | journal = Human Molecular Genetics | volume = 18 | issue = 6| pages = 1122–30 | doi = 10.1093/hmg/ddn427 | pmid = 19088127 | doi-access = free}}</ref> | |||
| <ref name="table">{{Cite journal|last1=Wen |first1=B. |last2=Li |first2=H. |last3=Lu |first3=D. |last4=Song |first4=X. |last5=Zhang |first5=F. |last6=He |first6=Y. |last7=Li |first7=F. |last8=Gao |first8=Y. |last9=Mao |first9=X. |last10=Zhang |first10=Liang |last11=Qian |first11=Ji |last12=Tan |first12=Jingze |last13=Jin |first13=Jianzhong |last14=Huang |first14=Wei |last15=Deka |first15=Ranjan |last16=Su |first16=Bing |last17=Chakraborty |first17=Ranajit |last18=Jin |first18=Li |title=Genetic evidence supports demic diffusion of Han culture |journal=Nature |volume=431 |issue=7006 |pages=302–05 |date=Sep 2004 |doi=10.1038/nature02878 |pmid=15372031 |url=http://159.226.149.45/compgenegroup/paper/wenbo%20Han%20culture%20paper%20(2004).pdf |display-authors=8 |url-status=dead |archive-url=https://web.archive.org/web/20090324201026/http://159.226.149.45/compgenegroup/paper/wenbo%20Han%20culture%20paper%20%282004%29.pdf |archive-date=24 March 2009 |bibcode=2004Natur.431..302W |s2cid=4301581}}</ref> | |||
| <ref name="Sato evab192">{{cite journal | vauthors = Sato T, Adachi N, Kimura R, Hosomichi K, Yoneda M, Oota H, Tajima A, Toyoda A, Kanzawa-Kiriyama H, Matsumae H, Koganebuchi K, Shimizu KK, Shinoda KI, Hanihara T, Weber A, Kato H, Ishida H | display-authors = 6 | title = Whole-Genome Sequencing of a 900-Year-Old Human Skeleton Supports Two Past Migration Events from the Russian Far East to Northern Japan | journal = Genome Biology and Evolution | volume = 13 | issue = 9 | pages = evab192 | date = September 2021 | pmid = 34410389 | pmc = 8449830 | doi = 10.1093/gbe/evab192 }}</ref> | |||
| <ref name=":0">{{Cite journal |vauthors=Zhao YB, Zhang Y, Zhang QC, Li HJ, Cui YQ, Xu Z, Jin L, Zhou H, Zhu H |date=2015 |title=Ancient DNA reveals that the genetic structure of the northern Han Chinese was shaped prior to 3,000 years ago |journal=PLOS ONE |volume=10 |issue=5 |pages=e0125676 |bibcode=2015PLoSO..1025676Z |doi=10.1371/journal.pone.0125676 |pmc=4418768 |pmid=25938511 |doi-access=free}}</ref> | |||
| <ref name=" |
<ref name="Wen2004">{{Cite journal |display-authors=6 |vauthors=Wen B, Li H, Lu D, Song X, Zhang F, He Y, Li F, Gao Y, Mao X, Zhang L, Qian J, Tan J, Jin J, Huang W, Deka R, Su B, Chakraborty R, Jin L |date=September 2004 |title=Genetic evidence supports demic diffusion of Han culture |journal=Nature |volume=431 |issue=7006 |pages=302–305 |bibcode=2004Natur.431..302W |doi=10.1038/nature02878 |pmid=15372031 |s2cid=4301581}}</ref> | ||
| <ref name=":3">{{Cite journal |last1=He |first1=Guanglin |last2=Wang |first2=Mengge |last3=Miao |first3=Lei |last4=Chen |first4=Jing |last5=Zhao |first5=Jie |last6=Sun |first6=Qiuxia |last7=Duan |first7=Shuhan |last8=Wang |first8=Zhiyong |last9=Xu |first9=Xiaofei |last10=Sun |first10=Yuntao |last11=Liu |first11=Yan |last12=Liu |first12=Jing |last13=Wang |first13=Zheng |last14=Wei |first14=Lanhai |last15=Liu |first15=Chao |date=28 March 2023 |title=Multiple founding paternal lineages inferred from the newly-developed 639-plex Y-SNP panel suggested the complex admixture and migration history of Chinese people |journal=Human Genomics |volume=17 |issue=1 |pages=29 |doi=10.1186/s40246-023-00476-6 |issn=1479-7364 |pmc=10045532 |pmid=36973821 |doi-access=free}}</ref> | |||
| <ref name=":5">{{Cite journal |display-authors=6 |vauthors=Damgaard PB, Marchi N, Rasmussen S, Peyrot M, Renaud G, Korneliussen T, Moreno-Mayar JV, Pedersen MW, Goldberg A, Usmanova E, Baimukhanov N, Loman V, Hedeager L, Pedersen AG, Nielsen K, Afanasiev G, Akmatov K, Aldashev A, Alpaslan A, Baimbetov G, Bazaliiskii VI, Beisenov A, Boldbaatar B, Boldgiv B, Dorzhu C, Ellingvag S, Erdenebaatar D, Dajani R, Dmitriev E, Evdokimov V, Frei KM, Gromov A, Goryachev A, Hakonarson H, Hegay T, Khachatryan Z, Khaskhanov R, Kitov E, Kolbina A, Kubatbek T, Kukushkin A, Kukushkin I, Lau N, Margaryan A, Merkyte I, Mertz IV, Mertz VK, Mijiddorj E, Moiyesev V, Mukhtarova G, Nurmukhanbetov B, Orozbekova Z, Panyushkina I, Pieta K, Smrčka V, Shevnina I, Logvin A, Sjögren KG, Štolcová T, Taravella AM, Tashbaeva K, Tkachev A, Tulegenov T, Voyakin D, Yepiskoposyan L, Undrakhbold S, Varfolomeev V, Weber A, Wilson Sayres MA, Kradin N, Allentoft ME, Orlando L, Nielsen R, Sikora M, Heyer E, Kristiansen K, Willerslev E |date=May 2018 |title=137 ancient human genomes from across the Eurasian steppes |journal=Nature |volume=557 |issue=7705 |pages=369–374 |bibcode=2018Natur.557..369D |doi=10.1038/s41586-018-0094-2 |pmid=29743675 |hdl=1887/3202709 |s2cid=13670282|hdl-access=free}}</ref> | |||
| <ref name="Yan2011">{{cite journal |vauthors=Yan S, Wang CC, Li H, Li SL, Jin L |date=September 2011 |title=An updated tree of Y-chromosome Haplogroup O and revised phylogenetic positions of mutations P164 and PK4 |journal=European Journal of Human Genetics |volume=19 |issue=9 |pages=1013–5 |doi=10.1038/ejhg.2011.64 |pmc=3179364 |pmid=21505448}}</ref> | |||
| <ref name="Moreno-Mayar2018">{{Cite journal |display-authors=6 |vauthors=Moreno-Mayar JV, Potter BA, Vinner L, Steinrücken M, Rasmussen S, Terhorst J, Kamm JA, Albrechtsen A, Malaspinas AS, Sikora M, Reuther JD, Irish JD, Malhi RS, Orlando L, Song YS, Nielsen R, Meltzer DJ, Willerslev E |date=January 2018 |title=Terminal Pleistocene Alaskan genome reveals first founding population of Native Americans |url=https://researchonline.ljmu.ac.uk/id/eprint/7887/1/UpwardSun_Nature%20paper%20MS%20DEC17.pdf |journal=Nature |volume=553 |issue=7687 |pages=203–207 |bibcode=2018Natur.553..203M |doi=10.1038/nature25173 |pmid=29323294 |s2cid=4454580}}</ref> | |||
| <ref name="ChaubeyEast">{{cite journal | vauthors = Chaubey G |title=East Asian ancestry in India |date=January 2015 |journal=Indian Journal of Physical Anthropology and Human Genetics |volume=34 |issue=2 |pages=193–199 |url= https://serialsjournals.com/abstract/78963_2.pdf |quote=Here the analysis of genome wide data on Indian and East/Southeast Asian demonstrated their restricted distinctive ancestry in India mainly running along the foothills of Himalaya and northeastern part.}}</ref> | |||
| <ref name="Chaubey_2010">{{cite journal | vauthors = Chaubey G, Metspalu M, Choi Y, Mägi R, Romero IG, Soares P, van Oven M, Behar DM, Rootsi S, Hudjashov G, Mallick CB, Karmin M, Nelis M, Parik J, Reddy AG, Metspalu E, van Driem G, Xue Y, Tyler-Smith C, Thangaraj K, Singh L, Remm M, Richards MB, Lahr MM, Kayser M, Villems R, Kivisild T | display-authors = 6 | title = Population genetic structure in Indian Austroasiatic speakers: the role of landscape barriers and sex-specific admixture | journal = Molecular Biology and Evolution | volume = 28 | issue = 2 | pages = 1013–1024 | date = February 2011 | pmid = 20978040 | pmc = 3355372 | doi = 10.1093/molbev/msq288}}</ref> | |||
| <ref name="Zhang_2015">{{Cite journal |vauthors=Zhang X, Liao S, Qi X, Liu J, Kampuansai J, Zhang H, Yang Z, Serey B, Sovannary T, Bunnath L, Seang Aun H, Samnom H, Kangwanpong D, Shi H, Su B |date=October 2015 |title=Y-chromosome diversity suggests southern origin and Paleolithic backwave migration of Austro-Asiatic speakers from eastern Asia to the Indian subcontinent |journal=Scientific Reports |volume=5 |pages=15486 |bibcode=2015NatSR...515486Z |doi=10.1038/srep15486 |pmc=4611482 |pmid=26482917}}</ref> | |||
| <ref name="Chaubey_2011">{{Cite journal |display-authors=6 |vauthors=Chaubey G, Metspalu M, Choi Y, Mägi R, Romero IG, Soares P, van Oven M, Behar DM, Rootsi S, Hudjashov G, Mallick CB, Karmin M, Nelis M, Parik J, Reddy AG, Metspalu E, van Driem G, Xue Y, Tyler-Smith C, Thangaraj K, Singh L, Remm M, Richards MB, Lahr MM, Kayser M, Villems R, Kivisild T |date=February 2011 |title=Population genetic structure in Indian Austroasiatic speakers: the role of landscape barriers and sex-specific admixture |journal=Molecular Biology and Evolution |volume=28 |issue=2 |pages=1013–1024 |doi=10.1093/molbev/msq288 |pmc=3355372 |pmid=20978040}}</ref> | |||
| <ref name="Van_Driem_2007">{{Cite journal |vauthors=Van Driem G |date=2007 |title=Austroasiatic phylogeny and the Austroasiatic homeland in light of recent population genetic studies. |url=https://www.academia.edu/download/36271840/2007MKS.pdf |journal=] |volume=37 |pages=1–14}}{{dead link|date=July 2022|bot=medic}}{{cbignore|bot=medic}}</ref> | |||
| <ref name="ArunKumar_2015">{{cite journal | vauthors = Arunkumar G, Wei LH, Kavitha VJ, Syama A, Arun VS, Sathua S, Sahoo R, Balakrishnan R, Riba T, Chakravarthy J, Chaudhury B, Bapukan P, Panda P, Das PK, Nayak PK, Li J, Pitchappan R | display-authors = 6 |title=A late Neolithic expansion of Y chromosomal haplogroup O2a1-M95 from east to west: Late Neolithic expansion of O2a1-M95 |journal=Journal of Systematics and Evolution |date=November 2015 |volume=53 |issue=6 |pages=546–560 |doi=10.1111/jse.12147 |s2cid=83103649 |language=en|doi-access=free}}</ref> | |||
| <ref name="Riccio2011">{{Cite journal |vauthors=Riccio ME, Nunes JM, Rahal M, Kervaire B, Tiercy JM, Sanchez-Mazas A |date=June 2011 |title=The Austroasiatic Munda population from India and Its enigmatic origin: a HLA diversity study |journal=Human Biology |volume=83 |issue=3 |pages=405–35 |doi=10.3378/027.083.0306 |pmid=21740156 |s2cid=39428816}}</ref> | |||
| <ref name="Ness-_2014">{{Cite book |title=The Global Prehistory of Human Migration |vauthors=Ness I |date=2014 |publisher=John Wiley & Sons |isbn=978-1-118-97058-4 |location=Chichester, West Sussex |page=265}}</ref> | |||
| }} | }} | ||
| Line 363: | Line 289: | ||
| {{refbegin}} | {{refbegin}} | ||
| * {{cite bioRxiv | vauthors = Lazaridis I, Belfer-Cohen A, Mallick S, Patterson N, Cheronet O, Rohland N, Bar-Oz G, Bar-Yosef O, Jakeli N, Kvavadze E, Lordkipanidze D, Matzkevich Z, Meshveliani T, Culleton BJ, Kennett DJ, Pinhasi R, Reich D | display-authors = 6 |title=Paleolithic DNA from the Caucasus reveals core of West Eurasian ancestry |date=21 September 2018 |biorxiv=10.1101/423079}} | * {{cite bioRxiv | vauthors = Lazaridis I, Belfer-Cohen A, Mallick S, Patterson N, Cheronet O, Rohland N, Bar-Oz G, Bar-Yosef O, Jakeli N, Kvavadze E, Lordkipanidze D, Matzkevich Z, Meshveliani T, Culleton BJ, Kennett DJ, Pinhasi R, Reich D | display-authors = 6 |title=Paleolithic DNA from the Caucasus reveals core of West Eurasian ancestry |date=21 September 2018 |biorxiv=10.1101/423079}} | ||
| * {{cite journal | vauthors = Lee J, Miller BK, Bayarsaikhan J, Johannesson E, Ventresca Miller A, Warinner C, Jeong C | title = Genetic population structure of the Xiongnu Empire at imperial and local scales | journal = Science Advances | volume = 9 | issue = 15 | pages = eadf3904 | date = April 2023 | pmid = 37058560 | doi = 10.1126/sciadv.adf3904 | ref = {{harvid|Lee|2023}} | doi-access = free }} | * {{cite journal | vauthors = Lee J, Miller BK, Bayarsaikhan J, Johannesson E, Ventresca Miller A, Warinner C, Jeong C | title = Genetic population structure of the Xiongnu Empire at imperial and local scales | journal = Science Advances | volume = 9 | issue = 15 | pages = eadf3904 | date = April 2023 | pmid = 37058560 | doi = 10.1126/sciadv.adf3904 | pmc = 10104459 | bibcode = 2023SciA....9F3904L | ref = {{harvid|Lee|2023}} | doi-access = free }} | ||
| * {{cite journal | vauthors = Li J, Zhang Y, Zhao Y, Chen Y, Ochir A, Zhu H, Zhou H | title = The genome of an ancient Rouran individual reveals an important paternal lineage in the Donghu population | journal = American Journal of Physical Anthropology | volume = 166 | issue = 4 | pages = 895–905 | date = August 2018 | pmid = 29681138 | doi = 10.1002/ajpa.23491 | ref = {{harvid|Li et al.|2018}} | publisher = ] }} | * {{cite journal | vauthors = Li J, Zhang Y, Zhao Y, Chen Y, Ochir A, Zhu H, Zhou H | title = The genome of an ancient Rouran individual reveals an important paternal lineage in the Donghu population | journal = American Journal of Physical Anthropology | volume = 166 | issue = 4 | pages = 895–905 | date = August 2018 | pmid = 29681138 | doi = 10.1002/ajpa.23491 | ref = {{harvid|Li et al.|2018}} | publisher = ] }} | ||
| {{refend}} | {{refend}} | ||
| Line 374: | Line 300: | ||
| {{Human genetics}} | {{Human genetics}} | ||
| ] | ] | ||
| ] | ] | ||
| ] | |||
| ] | |||
| ] | ] | ||
| ] | |||
| ] | |||
| ] | ] | ||
| ] | ] | ||
| ] | |||
| ] | |||
| ] | |||
Latest revision as of 12:42, 9 January 2025
Genetic history of East Asian peoplesThis article summarizes the genetic makeup and population history of East Asian peoples and their connection to genetically related populations such as Southeast Asians and North Asians, as well as Oceanians, and partly, Central Asians, South Asians, and Native Americans. They are collectively referred to as "East Eurasians" in population genomics.
Overview
-
 Phylogenetic position of East Asian lineages among other Eastern Eurasians.
Phylogenetic position of East Asian lineages among other Eastern Eurasians.
-
 Schematic of Populations in Eurasia from 45 to 10 kaBP.
Schematic of Populations in Eurasia from 45 to 10 kaBP.
-
 Highlighted regions show where ancient individuals associated with the labeled ancestry have been sampled.
Highlighted regions show where ancient individuals associated with the labeled ancestry have been sampled.

Population genomic research has studied the origin and formation of modern East Asians. The ancestors of East Asians (Ancient East Eurasians) split from other human populations possibly as early as 70,000 to 50,000 years ago. Possible routes into East Asia include a northern route model from Central Asia, beginning north of the Himalayas, and a southern route model, beginning south of the Himalayas and moving through Southeast Asia. Seguin-Orlando et al. (2014) stated that East Asians diverged from West Eurasians, which occurred at least 36,200 years ago, during the Upper Paleolithic. Vallini et al. 2024 noted that this divergence most likely occurred on the Persian Plateau 48,000 years ago.
Phylogenetic data suggests that an early Initial Upper Paleolithic wave (>45kya) "ascribed to a population movement with uniform genetic features and material culture" (Ancient East Eurasians) used a Southern dispersal route through South Asia, where they subsequently diverged rapidly, and gave rise to Australasians (Oceanians), the Ancient Ancestral South Indians (AASI), as well as Andamanese and East/Southeast Asians, although Papuans may have also received some geneflow from an earlier group (xOoA), around 2%, next to additional archaic admixture in the Sahul region.

The southern route model for East Asians has been corroborated in multiple recent studies, showing that most of the ancestry of Eastern Asians arrived from the southern route in to Southeast Asia at a very early period, starting perhaps as early as 70,000 years ago, and dispersed northward across Eastern Asia. However, genetic evidence also supports more recent migrations to East Asia from Central Asia and West Eurasia along the northern route, as shown by the presence of haplogroups Q and R, as well as Ancient North Eurasian ancestry.
The southern migration wave likely diversified after settling within East Asia, while the northern wave, which probably arrived from the Eurasian steppe, mixed with the southern wave, probably in Siberia.
A review paper by Melinda A. Yang (in 2022) described the East- and Southeast Asian lineage (ESEA); which is ancestral to modern East Asians, Southeast Asians, Polynesians, and Siberians, originated in Mainland Southeast Asia at c. 50,000 BCE, and expanded through multiple migration waves southwards and northwards, respectively. The ESEA lineage is also ancestral to the "basal Asian" Hoabinhian hunter-gatherers of Southeast Asia and the c. 40,000-year-old Tianyuan lineage found in Northern China, which can already be differentiated from the deeply related Ancestral Ancient South Indians (AASI) and Australasian (AA) lineages. There are currently eight detected, closely related, sub-ancestries in the ESEA lineage:
- Amur ancestry (ANA) – Associated with populations in the Amur River region, Mongolia, and Siberia, as well as parts of Central Asia.
- Fujian ancestry – Associated with ancient samples in the Fujian region of Southern China, and modern Austronesian-speaking populations.
- Guangxi ancestry – Associated with a 10,500-year-old individual from Longlin, Guangxi. This ancestry was not observed in either historical samples from Guangxi or contemporary East and Southeast Asians, suggesting that the lineage is extinct in the modern day.
- Jōmon ancestry – Ancestry associated with 8,000–3,000-year-old individuals in the Japanese archipelago.
- Hoabinhian ancestry – Ancestry on the ESEA lineage associated with 8,000–4,000-year-old hunter-gatherers in Laos and Malaysia.
- Tianyuan ancestry – Ancestry on the ESEA lineage associated with an Upper Paleolithic individual dating to 40,000 years ago in northern China.
- Tibetan ancestry – Associated with 3,000-year-old individuals in the Himalayan region of the Tibetan Plateau.
- Yellow River ancestry – Associated with populations in the Yellow River region and common among Sino-Tibetan-speakers.
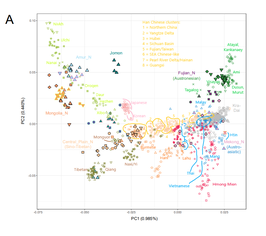
The genetic makeup of East Asians is primarily characterized by "Yellow River" (East Asian) ancestry which formed from a major Ancient Northern East Asian (ANEA) component and a minor Ancient Southern East Asian (ASEA) one. The two lineages diverged from each other at least 19,000 years ago, after the divergence of the Jōmon, Guangxi (Longlin), Hoabinhian and Tianyuan lineages. Contemporary East Asians (notably Sino-Tibetan speakers) mostly have Yellow River ancestry, which is associated with millet and rice cultivation. "East Asian Highlanders" (Tibetans) carry both Tibetan ancestry and Yellow River ancestry. Japanese people were found to have a tripartite origin; consisting of Jōmon ancestry, Amur ancestry, and Yellow River ancestry. East Asians carry a variation of the MFSD12 gene, which is responsible for lighter skin colour. Huang et al. (2021) found evidence for light skin being selected among the ancestral populations of West Eurasians and East Eurasians, prior to their divergence.
Northeast Asians such as Tungusic, Mongolic, and Turkic peoples derive most of their ancestry from the "Amur" (Ancient Northeast Asian) subgroup of the Ancient Northern East Asians, which expanded massively with millet cultivation and pastoralism. Tungusic peoples display the highest genetic affinity to Ancient Northeast Asians, represented by c. 7,000 and 13,000 year old specimens, whereas Turkic peoples have significant West Eurasian admixture.
East Asian populations exhibit some European-related admixture, originating from Silk Road traders and interactions with Mongolians, who were well-acquainted with European-like populations. This is more common among northern Han Chinese (2.8%) than southern Han Chinese (1.7%), Japanese (2.2%), and Koreans (1.6%). However, East Asians have less European-related admixture than Northeast Asians like Mongolians (10.9%), Oroqen (9.6%), Daur (8.0%), and Hezhen (6.8%).
Austronesians mainly carry "Fujian" (Ancient Southern East Asian) ancestry, which is associated with the spread of rice cultivation. Isolated hunter-gatherers in Southeast Asia, specifically in Malaysia and Thailand, such as the Semang, derive most of their ancestry from the Hoabinhian lineage. The emergence of the Neolithic in Southeast Asia went along with a population shift caused by migrations from southern China. Neolithic Mainland Southeast Asian samples predominantly have Ancient Southern East Asian ancestry with Hoabinhian-related admixture. In modern populations, this admixture of Ancient Southern East Asian and Hoabinhian ancestry is most strongly associated with Austroasiatic speakers.
An early branch of Ancient Northern East Asians, together with Ancient North Eurasians, gave rise to the Ancient Paleo-Siberians, who in turn gave rise to both "modern Paleo-Siberians" (such as Chukotko-Kamchatkan, Yeniseian, and Nivkh speakers) and contemporary Native Americans. Paleo-Siberian ancestry was once widespread across North Asia, but it was largely replaced by Neo-Siberian ancestry due to a major population turnover from the south, possibly involving Uralic and Yukaghir speakers. This was later followed by another expansion from the south in relatively recent times, associated with Amur River ancestry involving Tungusic, Mongolic, and Turkic speakers.
Ancient and historical populations
Xiongnu people
Main article: XiongnuThe Xiongnu, possibly a Turkic, Mongolic, Yeniseian or multi-ethnic people, were a confederation of nomadic peoples who, according to ancient Chinese sources, inhabited the eastern Eurasian Steppe from the 3rd century BC to the late 1st century AD. Chinese sources report that Modu Chanyu, the supreme leader after 209 BC, founded the Xiongnu Empire.
Autosomal DNA
It was found that the "predominant part of the Xiongnu population is likely to have spoken Turkic". However, important cultural, technological and political elements may have been transmitted by Eastern Iranian-speaking Steppe nomads: "Arguably, these Iranian-speaking groups were assimilated over time by the predominant Turkic-speaking part of the Xiongnu population". This is reflected by the average genetic makeup of Xiongnu samples, having approximately 58% East Eurasian ancestry, represented by a Bronze Age population from Khövsgöl, Mongolia, which may be associated with the Turkic linguistic heritage. The rest of the Xiongnu's ancestry (~40%) was related to West Eurasians, represented by the Gonur Depe BMAC population of Central Asia, and the Sintashta culture of the Western steppe. The Xiongnu displayed striking heterogeneity and could be differentiated into two subgroups, "Western Xiongnu" and "Eastern Xiongnu", with the former being of "hybrid" origins displaying affinity to previous Saka tribes, such as represented by the Chandman culture, while the later was of primarily Ancient Northeast Asian (Ulaanzuukh-Slab Grave) origin. High status Xiongnu individuals tended to have less genetic diversity, and their ancestry was essentially derived from the Eastern Eurasian Ulaanzuukh/Slab Grave culture.
Paternal lineages
A review of the available research has shown that, as a whole, 53% of Xiongnu paternal haplogroups were East Eurasian, while 47% were West Eurasian. In 2012, Chinese researchers published an analysis of the paternal haplogroups of 12 elite Xiongnu male specimens from Heigouliang in Xinjiang, China. Six of the specimens belonged to Q1a, while four belonged to Q1b-M378. 2 belonged to unidentified clades of Q*. In another study, a probable Chanyu of the Xiongnu empire was assigned to haplogroup R1.
Maternal lineages
The bulk of the genetics research indicates that, as a whole, 73% of Xiongnu maternal haplogroups were East Eurasian, while 27% were West Eurasian. A 2003 study found that 89% of Xiongnu maternal lineages from the Egiin Gol valley were of East Asian origin, while 11% were of West Eurasian origin. A 2016 study of Xiongnu from central Mongolia found a considerably higher frequency of West Eurasian maternal lineages, at 37.5%.
Xianbei people
Main article: XianbeiAutosomal DNA
A full genome study on multiple Xianbei remains found them to be derived primarily to exclusively from the Ancient Northeast Asian gene pool.
Paternal lineages
A genetic study published in the American Journal of Physical Anthropology in August 2018 noted that the paternal haplogroup C2b1a1b has been detected among the Xianbei and the Rouran, and was probably an important lineage among the Donghu people.
Maternal lineages
Genetic studies published in 2006 and 2015 revealed that the mitochondrial haplogroups of Xianbei remains were of East Asian origin. According to Zhou (2006) the maternal haplogroup frequencies of the Tuoba Xianbei were 43.75% haplogroup D, 31.25% haplogroup C, 12.5% haplogroup B, 6.25% haplogroup A and 6.25% "other". Zhou (2014) obtained mitochondrial DNA analysis from 17 Tuoba Xianbei, which indicated that these specimens were, similarly, completely East Asian in their maternal origins, belonging to haplogroups D, C, B, A, O and haplogroup G.
Jōmon people
Main article: Jōmon peopleThe Jōmon people represent the indigenous population of the Japanese archipelago during the Jōmon period. They are inferred to descend from the Paleolithic inhabitants of Japan. Genetic analyses on Jōmon remains found them to represent a deeply diverged East Asian lineage. The Jōmon lineage is inferred to have diverged from Ancient East Asians before the divergence between Ancient Northern East Asians and Ancient Southern East Asians, but after the divergence of the basal Tianyuan man and or Hoabinhians. Beyond their broad affinity with Eastern Asian lineages, the Jōmon also display a weak affinity for Ancient North Eurasians (ANE), which may be associated with the introduction of microblade technology to Northeast Asia and northern East Asia during the Last Glacial Maximum via the ANE or Ancient Paleo-Siberians.
Hoabinhians
Main article: HoabinhianThe Hoabinhians represent a technologically advanced society of hunter-gatherers, primarily living in Mainland Southeast Asia, but also adjacent regions of Southern China. While the Upper Paleolithic origins of this 'Hoabinhian ancestry' are unknown, Hoabinhian ancestry has been found to be related to the main 'East Asian' ancestry component found in most modern East and Southeast Asians, although deeply diverged from it. Together with the Paleolithic Tianyuan man, they form early branches of East Asian genetic diversity, and are described as "Basal Asian" (BA) or "Basal East Asian" (BEA).
Modern populations
Manchu and Daur peoples
Main article: Tungusic peoplesAutosomal DNA
A study on the Manchu population of Liaoning reported that they have a close genetic relationship and significant admixture signals from northern Han Chinese. The Liaoning Manchu were formed from a major ancestral component related to Yellow River farmers and a minor ancestral component linked to ancient populations from the Amur River Basin, or others. The Manchu were therefore an exception to the coherent genetic structure of Tungusic-speaking populations, likely due to the large-scale population migrations and genetic admixtures in the past few hundred years.
Paternal lineages
A plurality of Daur males belong to Haplogroup C-M217 (12/39 = 30.8% according to Xue Yali et al. 2006, 88/207 = 42.5% according to Wang Chi-zao et al. 2018), with Haplogroup O-M122 being the second most common haplogroup among present-day Daurs (10/39 = 25.6%, 52/207 = 25.1%). There are also tribes (hala; cf. Kazakh tribes) among the Daurs that belong predominantly to other Y-DNA haplogroups, such as Haplogroup N-M46/M178 (Merden hala) and Haplogroup O1b1a1a-M95 (Gobulo hala). Haplogroup C3b2b1*-M401(xF5483) has been identified as a possible marker of the Aisin Gioro and is found in ten different ethnic minorities in northern China, but is less prevalent from Han Chinese. The Manchu people also display a significant amount of haplogroup C-M217, but the most often observed Y-DNA haplogroup among present-day Manchus is Haplogroup O-M122, which they share in common with the general population of China.
Ainu people
Main article: Ainu peopleThe exact origins of the early Ainu remains unclear, but it is generally agreed to be linked to the Satsumon culture of the Epi-Jōmon period, with later influences from the nearby Okhotsk culture. The Ainu appear genetically most closely related to the Jōmon period peoples of Japan. The genetic makeup of the Ainu represents a "deep branch of East Asian diversity". Compared to contemporary East Asian populations, the Ainu share "a closer genetic relationship with northeast Siberians".
Japanese people
Main articles: Japanese people and Genetic and anthropometric studies on Japanese people
Japanese populations in modern Japan can be traced to three separate, but related demographics: the Ainu, Ryukyuan and Mainland Japanese (Yamato). The populations are closely related to clusters found in Northeastern Asia with the Ainu group being most similar to the Ryukyuan group, the Ryukyuan group being most similar to the Yamato group, and the Yamato group being most similar to Koreans among other East Asian people.
Autosomal DNA
The majority of Japanese genetic ancestry is derived from sources related to other mainland Asian groups, mostly Koreans, while the other amount is derived from the local Jōmon hunter-gatherers.
According to a full genome analyses, the modern Japanese harbor a Northeast Asian (Amur River ancestry), an East Asian (Yellow River ancestry), and an indigenous Jōmon component. In addition to the indigenous Jōmon hunter-gatherers and the Yayoi period migrants, a new strand was hypothesized to have been introduced during the Yayoi-Kofun transition period that had strong cultural and political affinity with Korea and China. However, this theory is mildly criticized in recent years (see Genetic and anthropometric studies on Japanese people#Tripartite ancestry theory).
Paternal lineages
A comprehensive study of worldwide Y-DNA diversity (Underhill et al. 2000) included a sample of 23 males from Japan, of whom 35% belonged to haplogroup D-M174, 26% belonged to O-M175, 22% belonged to O-M122, 13% belonged to C-M8 and C-M130, and 4.3% belonged to N-M128. Poznik et al. (2016) reported the haplogroups of a sample of Japanese men from Tokyo: 36% belonged to D2-M179, 32% had O2b-M176, 18% carried O3-M122, 7.1% carried C1a1-M8, 3.6% belonged to O2a-K18, and 3.6% carried C2-M217.
Maternal lineages
According to an analysis of the 1000 Genomes Project's sample of Japanese collected in the Tokyo metropolitan area, the mtDNA haplogroups found among modern Japanese include D (35.6%), B (13.6%), M7 (10.2%), G (10.2%), N9 (8.5%), F (7.6%), A (6.8%), Z (3.4%), M9 (2.5%), and M8 (1.7%).
Korean people
Main article: KoreansKorean populations in modern Korea can be traced to many origins from the people of the Mumun period to the Yemaek people. In modern times, Koreans are related to other populations found in Northeast Asia, however according to recent studies, ancient Koreans included populations related to the Yayoi people, Jōmon people, Siberian influx and etc.
Autosomal DNA
Ancient genome comparisons revealed that the genetic makeup of Koreans can be best described as an admixture between Northeast Asian hunter-gatherers and an influx of rice-farming Southeast Asian agriculturalists from the Yangtze river valley. This is supported by archaeological, historical and linguistic evidence, which suggest that the direct ancestors of Koreans were proto-Koreans who inhabited the northeastern region of China and the Korean Peninsula during the Neolithic (8,000–1,000 BC) and Bronze (1,500–400 BC) Ages.
There is evidence for considerable genetic diversity, including elevated levels of Jōmon ancestry among early southern Koreans. It was hypothesized that the Jōmon ancestry of ancient Koreans was lost over time, as they continually mixed with incoming populations from northern China, followed by a period of isolation during the Three Kingdoms period, resulting in the homogenous gene pool of modern Koreans. A 2022 study was unable to detect significant Jōmon ancestry in modern Koreans, however by using different proxies of ancestry, a Jōmon contribution of 3.1–4.4% was found for present-day Ulsan Koreans. Nevertheless, the authors suggested that the model that yielded this result is not the most reliable.
Evidence for both Southern and Northern mtDNA and Y-DNA haplogroups has been observed in Koreans, similar to Japanese.
Over 70% of extant genetic diversity among Koreans can be explained by admixture with ancient South Chinese immigrants, who were related to Iron Age Cambodians.
Paternal lineages
Studies of polymorphisms in the human Y-chromosome have so far produced evidence to suggest that the Korean people have a long history as a distinct, mostly endogamous ethnic group, with successive waves of people moving to the peninsula and three major Y-chromosome haplogroups. A majority of Koreans belong to subclades of haplogroup O-M175 (ca. 79% in total, with about 42% to 44% belonging to haplogroup O2-M122, about 31% to 32% belonging to haplogroup O1b2-M176, and about 2% to 3% belonging to haplogroup O1a-M119), while a significant minority belong to subclades of haplogroup C2-M217 (ca. 12% to 13% in total). Other Y-DNA haplogroups, including haplogroup N-M231, haplogroup D-M55, and haplogroup Q-M242, are also found in smaller proportions of present-day Koreans.
Maternal lineages
Studies of Korean mitochondrial DNA lineages have shown that there is a high frequency of Haplogroup D4, followed by haplogroup B, and then haplogroup A and haplogroup G. Haplogroups with lower frequency include N9, Y, F, D5, M7, M8, M9, M10, M11, R11, C, and Z.
Mongolic peoples
Main article: Mongolic peoplesThe ethnogenesis of Mongolic peoples is largely linked with the expansion of Ancient Northeast Asians. They subsequently came into contact with other groups, notably Sinitic peoples to their South and Western Steppe Herders to their far West. The Mongolians pastoralist lifestyle, may in part be derived from the Western Steppe Herders, but without much geneflow between these two groups, suggesting cultural transmission. The Mongols are believed to be the descendants of the Xianbei and the proto-Mongols. The former term includes the Mongols proper (also known as the Khalkha Mongols), Oirats, the Kalmyk people and the Southern Mongols. The latter comprises the Abaga Mongols, Abaganar, Aohans, Baarins, Gorlos Mongols, Jalaids, Jaruud, Khishigten, Khuuchid, Muumyangan and Onnigud. The Daur people are descendants of the para-Mongolic Khitan people.
Paternal lineages
The majority of Mongols in Mongolia and Russia belong to subclades of haplogroup C-M217, followed by lower frequency of O-M175 and N-M231. A minority belongs to haplogroup Q-M242, and a variety of West Eurasian haplogroups.
Maternal lineages
The maternal haplogroups are diverse but similar to other northern Asian populations, including Haplogroup D, Haplogroup C, Haplogroup B, and Haplogroup A, which are shared among indigenous American and Asian populations. West Eurasian mtDNA haplogroups makes up a some minority percentages. Haplogroup HV, Haplogroup U, Haplogroup K, Haplogroup I, Haplogroup J are all found in Mongolic people.
Han Chinese
See also: Han Chinese
The origins of the Han Chinese are primarily from Neolithic Yellow River farmers, which formed from Ancient Northern East Asians (ANEA) and Neolithic groups near the Yangtze, which formed Ancient Southern East Asians (ASEA). Today's modern Han Chinese can be categorized out of convenience into two subgroups, Northern and Southern Han Chinese, despite there being no clear genetic divide between the north and south because the Han Chinese are a clinal population. The Northern Han Chinese carry mostly ANEA ancestry with some degree of ASEA admixture, whereas the Southern Han Chinese carry significantly higher levels of ASEA ancestry than Northern Han Chinese, although ANEA ancestry is still significant. In large, the Han Chinese cluster retains a level of singularity with its admixture of ANEA and ASEA ancestries which is unique to the group. When comparing with other East Asian populations, the Northern Han Chinese cluster is related to the "Korean/Yamato cluster" in terms of a correlative genetic relationship (mostly due to the overlap of ANEA), but is also quite distinguishable from them genetically, due to the presence of ASEA ancestry and the absence of Jōmon ancestry. The Southern Han Chinese also share more alleles with Thai and other Kra–Dai peoples according to principal component analysis than Northern Han Chinese.
The genetic makeup of the modern Han Chinese is not purely uniform in terms of physical appearance and biological structure due to the vast geographical expanse of China and the migratory percolations that have occurred throughout it over the last few millennia. This has also engendered the emergence and evolution of the diverse multiplicity of assorted Han subgroups found throughout the various regions of modern China today. Comparisons between the Y chromosome single-nucleotide polymorphisms (SNPs) and mitochondrial DNA (mtDNA) of modern Northern Han Chinese and 3000 year old Hengbei ancient samples from China's Central Plains show that they are extremely similar to each other. These findings demonstrate that the core fundamental structural basis that shaped the genetic makeup of the present-day Northern Han Chinese was already formed more than three thousand years ago.
Studies of DNA remnants from the Central Plains area of China 3000 years ago show close affinity between that population and those of Northern Han today in both the Y-DNA and mtDNA. Both northern and southern Han show similar Y-DNA genetic structure.
Northern Han Chinese populations also have some West Eurasian admixture, especially Han Chinese populations in Shaanxi (~2%-4.6%) and Liaoning (~2%). During the Zhou dynasty, or earlier, peoples with paternal haplogroup Q-M120 also contributed to the ethnogenesis of Han Chinese people. This haplogroup is implied to be widespread in the Eurasian steppe and north Asia since it is found among Cimmerians in Moldova and Bronze Age natives of Khövsgöl. But it is currently near-absent in these regions except for East Asia. In modern China, haplogroup Q-M120 can be found in the northern and eastern regions. Other Y-DNA haplogroups that have been found with notable frequency in samples of Han Chinese include O-P203 (15/165 = 9.1%, 217/2091 = 10.38%, 47/361 = 13.0%), C-M217 (10/168 = 6.0%, 27/361 = 7.5%, 176/2091 = 8.42%, 187/1730 = 10.8%, 20/166 = 12.0%), N-M231 (6/166 = 3.6%, 94/2091 = 4.50%, 18/361 = 5.0%, 117/1729 = 6.8%, 17/165 = 10.3%), O-M268(xM95, M176) (78/2091 = 3.73%, 54/1147 = 4.7%, 8/168 = 4.8%, 23/361 = 6.4%, 12/166 = 7.2%), and Q-M242 (2/168 = 1.2%, 49/1729 = 2.8%, 61/2091 = 2.92%, 12/361 = 3.3%, 48/1147 = 4.2%).
However, the mtDNA of Han Chinese increases in diversity as one looks from northern to southern China, which suggests that the influx of male Han Chinese migrants intermarried with the local female non-Han aborigines after arriving in what is now modern-day Guangdong, Fujian, and other regions of southern China. Despite this, tests comparing the genetic profiles of northern Han, southern Han, and non-Han southern natives determined that haplogroups O1b-M110, O2a1-M88 and O3d-M7, which are prevalent in non-Han southern natives, were only observed in some southern Han Chinese (4% on average), but not in the northern Han genetic profile. Therefore, this proves that the male contribution of the southern non-Han natives in the southern Han genetic profile is limited, assuming that the frequency distribution of Y lineages in southern non-Han natives represents that prior to the expansion of Han culture which originated two thousand years ago from the north.
A recent, and to date the most extensive, genome-wide association study of the Han population, shows that geographic-genetic stratification from north to south has occurred and centrally placed populations act as the conduit for outlying ones. Ultimately, with the exception in some ethnolinguistic branches of the Han Chinese, such as Pinghua and Tanka people, there is a "coherent genetic structure" found in the entirety of the modern Han Chinese populace. Although admixture proportions can vary according to geographic region, the average genetic distance between various Han Chinese populations is much lower than between European populations, for example.
Autosomal DNA
A 2018 study calculated pairwise FST (a measure of genetic difference) based on genome-wide SNPs, among the Han Chinese (Northern Han from Beijing and Southern Han from Hunan, Jiangsu and Fujian provinces), Japanese and Korean populations sampled. It found that the smallest FST value was between Northern Han Chinese (Beijing) (CHB) and Southern Han (Hunan, Fujian, etc.) Chinese (CHS) (FST = 0.0014), while CHB and Korean (KOR) (FST = 0.0026) and between KOR and Japanese (JPT) (FST = 0.0033). Generally, pairwise FST between Han Chinese, Japanese and Korean (0.0026~ 0.0090) are greater than that within Han Chinese (0.0014). These results suggested Han Chinese, Japanese and Korean are different in terms of genetic make-up, and the differences among the three groups are much larger than that between northern and southern Han Chinese. Nonetheless, there is also genetic diversity among the Southern Han Chinese. The genetic composition of the Han population in Fujian might not accurately represent that of the Han population in Guangdong.
Another study shows that the northern and southern Han Chinese are genetically close to each other and it finds that the genetic characteristics of present-day northern Han Chinese were already formed prior to three thousand years ago in the Central Plain area.
A recent genetic study on the remains of people (~4,000 years BP) from the Mogou site in the Gansu-Qinghai (or Ganqing) region of China revealed more information on the genetic contributions of these ancient Di-Qiang people to the ancestors of the Northern Han. It was deduced that 3,300 to 3,800 years ago some Mogou people had merged into the ancestral Han population, resulting in the Mogou people being similar to some northern Han in sharing up to ~33% paternal (O3a) and ~70% maternal (D, A, F, M10) haplogroups. The mixing ratio was possibly 13–18%.
The estimated contribution of northern Han to southern Han is substantial in both paternal and maternal lineages and a geographic cline exists for mtDNA. As a result, the northern Han are one of the primary contributors to the gene pool of the southern Han. However, it is noteworthy that the expansion process was not only dominated by males, as is shown by both contribution of the Y-chromosome and the mtDNA from northern Han to southern Han. Northern Han Chinese and Southern Han Chinese exhibit both Ancient Northern East Asian and Ancient Southern East Asian ancestries. These genetic observations are in line with historical records of continuous and large migratory waves of northern China inhabitants escaping warfare and famine, to southern China. Aside from these large migratory waves, other smaller southward migrations occurred during almost all periods in the past two millennia. A study by the Chinese Academy of Sciences into the gene frequency data of Han subpopulations and ethnic minorities in China showed that Han subpopulations in different regions are also genetically quite close to the local ethnic minorities, suggesting that in many cases, ethnic minorities ancestry had mixed with Han, while at the same time, the Han ancestry had also mixed with the local ethnic minorities.
Han Chinese, similar to other East Asian populations, have inherited West Eurasian ancestry, around 2.8% in Northern Han Chinese and around 1.7% in Southern Han Chinese.
An extensive, genome-wide association study of the Han population in 2008, shows that geographic-genetic stratification from north to south has occurred and centrally placed populations act as the conduit for outlying ones. Ultimately, with the exception in some ethnolinguistic branches of the Han Chinese, such as Pinghua, there is "coherent genetic structure" (homogeneity) in all Han Chinese.
Paternal lineages
The major haplogroups of Han Chinese belong to subclades of Haplogroup O-M175. Y-chromosome O2-M122 is a common DNA marker in Han Chinese, as it appeared in China in prehistoric times, and is found in approximately 50% of Chinese males, with frequencies tending to be high toward the east of the country, ranging from 29.7% to 52% in Han from southern and central China, to 55–68% in Han from the eastern and northeastern Chinese mainland and Taiwan.
Other Y-DNA haplogroups that have been found with notable frequency in samples of Han Chinese include O-P203 (9.1–13.0%), C-M217 (6.0–12.0%), N-M231 (3.6–10.3%), O-M268(xM95, M176) (4.7–7%), and Q-M242 (2/168 = 1.2–4.2%).
Maternal lineages
The mitochondrial-DNA haplogroups of the Han Chinese can be classified into the northern East Asian-dominating haplogroups, including A, C, D, G, M8, M9, and Z, and the southern East Asian-dominating haplogroups, including B, F, M7, N*, and R.
These haplogroups account for 52.7% and 33.85% of those in the Northern Han, respectively. Haplogroup D is the modal mtDNA haplogroup among northern East Asians. Among these haplogroups, D, B, F, and A were predominant in the Northern Han, with frequencies of 25.77%, 11.54%, 11.54%, and 8.08%, respectively.
However, in the Southern Han, the northern and southern East Asian-dominating mtDNA haplogroups accounted for 35.62% and 51.91%, respectively. The frequencies of haplogroups D, B, F, and A reached 15.68%, 20.85%, 16.29%, and 5.63%, respectively.
Tibetan peoples
Main article: Tibetan peopleThe ethnic roots of Tibetans can be traced back to a deep Eastern Asian lineage representing the indigenous population of the Tibetan plateau since c. 40,000 to 30,000 years ago, and arriving Neolithic farmers from the Yellow River within the last 10,000 years associated, and which can be associated with having introduced the Sino-Tibetan languages. Modern Tibetans derive up to 20% from Paleolithic Tibetans, with the remaining 80% being primarily derived from Yellow River farmers. The present-day Tibetan gene pool was formed at least 5,100 years BP.
Paternal lineage
Tibetan males predominantly belong to the paternal lineage D-M174 followed by lower amounts of O-M175.
Maternal lineage
Tibetan females belong mainly to the Northeast Asian maternal haplogroups M9a1a, M9a1b, D4g2, D4i and G2ac, showing continuity with ancient middle and upper Yellow River populations.
Turkic peoples
See also: Turkic peoplesLinguistic and genetic evidence strongly suggests an early presence of Turkic peoples in eastern Mongolia. The genetic evidence suggests that the Turkification of Central Asia was carried out by East Asian dominant minorities migrating out of Mongolia.
Genetic data found that almost all modern Turkic-speaking peoples retained at least some shared ancestry associated with "Southern Siberian and Mongolian" (SSM) populations, supporting this region as the "Inner Asian Homeland (IAH) of the pioneer carriers of Turkic languages" which subsequently expanded into Central Asia.


An Ancient Northeast Asian origin of the early Turkic peoples has been corroborated in multiple recent studies. Early and medieval Turkic groups however exhibited a wide range of both (Northern) East Asian and West Eurasian genetic origins, in part through long-term contact with neighboring peoples such as Iranian, Mongolic, Tocharian, Uralic and Yeniseian peoples, and others.
Paternal lineages
Common Y-DNA haplogroups in Turkic peoples are Haplogroup N-M231 (found with especially high frequency among Turkic peoples living in present-day Russia, especially among Siberian Tatars, as Zabolotnie Tatars have one of the highest frequencies of this haplogroup, second only to Samoyedic Nganasans ), Haplogroup C-M217 (especially in Central Asia, and in particular, Kazakhstan, also in Siberia among Siberian Tatars), Haplogroup Q-M242 (especially in Southern Siberia among the Siberian Tatars, also quite frequent among Lipka Tatars and among Turkmens and the Qangly tribe of Kazakhs), and Haplogroup O-M175 (especially among Turkic peoples living in present-day China, the Naiman tribe of Kazakhs and Siberian Tatars). Some groups also have Haplogroup R1b (notably frequent among the Teleuts, Siberian Tatars, and Kumandins of Southern Siberia, the Bashkirs of the Southern Ural region of Russia, and the Qypshaq tribe of Kazakhs), Haplogroup R1a (notably frequent among the Kyrgyz, Altaians, Siberian Tatars, Lipka Tatars, Volga Tatars, Crimean Tatars and several other Turkic peoples living in present-day Russia), Haplogroup J-M172 (especially frequent among Uyghurs, Azerbaijanis, and Turkish people), and Haplogroup D-M174 (especially among Yugurs, but also observed regularly with low frequency among Southern Altaians, Nogais, Kazakhs, and Uzbeks).
Relationship to other Asia-Pacific and Native American populations
Central Asians
See also: History of Central Asia § Medieval
The genetic evidence suggests that the Turkification of Central Asia was carried out by East Asian dominant minorities migrating out of Mongolia. According to a recent study, the Turkic Central Asian populations, such as Kyrgyz, Kazakhs, Uzbeks, and Turkmens share more of their gene pool with various East Asian and Siberian populations than with West Asian or European populations. The study further suggests that both migration and linguistic assimilation helped to spread the Turkic languages in Eurasia.
North Asians and Native Americans
Genetic data suggests that North Asia was populated during the Terminal Upper-Paleolithic (36±1.5ka) period from a distinct Paleolithic population migrating through Central Asia into Northern Siberia. This population is known as Ancient North Eurasians or Ancient North Siberians, who were of West Eurasian origin.
Between 30,000 and 25,000 years ago, the ancestors of both Paleo-Siberians and Native Americans originated from admixture between Ancient North Eurasians/Siberians and an Ancient East Asian lineage. Ancestral Native Americans (or Ancient Beringians) later migrated towards the Beringian region, became isolated from other populations, and subsequently populated the Americas. Further geneflow from Northeast Asia resulted in the modern distribution of "Neo-Siberians" (associated with Tungusic, Mongolic, and Turkic speakers) through the merger of Paleo-Siberians with Northeast Asians.
A study found that the North Asian ethnic groups—Altai Kazakh, Khanty, Komi (Zyrian), Mongols, Buryats, Dukha, and Yakuts—are, on average, with the exception of the Komi, more closely related to East Asians than to Europeans, but still occupy a distinct position from the major East Asian populations (typified by Koreans, Japanese, and Han Chinese), who formed a very tight cluster. "Analyses of all 122 populations confirm many known relationships and show that most populations from North Asia form a cluster distinct from all other groups. Refinement of analyses on smaller subsets of populations reinforces the distinctiveness of North Asia and shows that the North Asia cluster identifies a region that is ancestral to Native Americans."
Native Americans
Multiple studies suggests that all Native Americans ultimately descended from a single founding population that initially diverged from" Ancestral Beringians" which shared a common origin with Paleo-Siberians from the merger of Ancient North Eurasians and a Basal-East Asian source population in Mainland Southeast Asia around 36,000 years ago, at the same time at which the proper Jōmon people split from Basal-East Asians, either together or during a separate expansion wave. The basal northern and southern Native American branches, to which all other Indigenous peoples belong, diverged around 16,000 years ago, although earlier dates were also proposed. An indigenous American sample from 16,000 BCE in Idaho, which is craniometrically similar to modern Native Americans, was found to have been closely related to Paleosiberians, confirming that Ancestral Native Americans split from an ancient Siberian source population somewhere in northeastern Siberia. Genetic data on samples with alleged "Paleo-Indian" morphology turned out to be closely related to contemporary Native Americans, disproving a hypothetical earlier migration into the Americas. The scientists suggest that variation within Native American morphology is just that, the natural variation which have arisen during the formation of Ancestral Native Americans. Signals of a hypothetical "population Y", if not a false positive, are likely explained through a now extinct population from East Asia (e.g. Tianyuan man, which contributed low amounts of ancestry to the Ancestral Native American gene pool in Asia, and perhaps also towards other Asian and Oceanian populations.
South Asians
Main article: Genetic history of South AsiaThe genetic makeup of modern South Asians can be described as a combination of West Eurasian ancestries with divergent East Eurasian ancestries. The latter primarily include an indigenous South Asian component (termed Ancient Ancestral South Indians, short "AASI") that is distantly related to the Andamanese peoples, as well as to East Asians and Aboriginal Australians, and further include additional, regionally variable East/Southeast Asian components. The East Asian-related ancestry component forms the major ancestry among Tibeto-Burmese and Khasi-Aslian speakers in the Himalayan foothills and Northeast India, and is generally distributed throughout South Asia at lower frequency, with substantial presence in Mundari-speaking groups.
According to a genetic research (2015) including linguistic analyses, suggests an East Asian origin for proto-Austroasiatic groups, which first migrated to Southeast Asia and later into India. According to Ness, there are three broad theories on the origins of the Austroasiatic speakers, namely northeastern India, central or southern China, or southeast Asia. Multiple researches indicate that the Austroasiatic populations in India are derived from (mostly male dominated) migrations from Southeast Asia during the Holocene. According to Van Driem (2007), "...the mitochondrial picture indicates that the Munda maternal lineage derives from the earliest human settlers on the Subcontinent, whilst the predominant Y chromosome haplogroup argues for a Southeast Asian paternal homeland for Austroasiatic language communities in India."
According to Chaubey et al. (2011), "Austroasiatic speakers in India today are derived from dispersal from Southeast Asia, followed by extensive sex-specific admixture with local Indian populations." According to Zhang et al. (2015), Austroasiatic (male) migrations from southeast Asia into India took place after the lates Glacial maximum, circa 4,000 years ago. According to Arunkumar et al. (2015), Y-chromosomal haplogroup O2a1-M95, which is typical for Austroasiatic speaking peoples, clearly decreases from Laos to east India, with "a serial decrease in expansion time from east to west," namely "5.7 ± 0.3 Kya in Laos, 5.2 ± 0.6 in Northeast India, and 4.3 ± 0.2 in East India." This suggests "a late Neolithic east to west spread of the lineage O2a1-M95 from Laos." According to Riccio et al. (2011), the Munda people are likely descended from Austroasiatic migrants from southeast Asia. According to Ness, the Khasi probably migrated into India in the first millennium BCE.
According to Yelmen et al. 2019, the two main components of Indian genetic variation; the South Asian populations that "separated from East Asian and Andamanese populations" form one of the deepest splits among non-African groups compared to the West Eurasian component because of "40,000 years of independent evolution".
Geneflow from Southeast Asians (particularly Austroasiatic groups) to South Asian peoples is associated with the introduction of rice-agriculture to South Asia. There is significant cultural, linguistic, and political Austroasiatic influence on early India, which can also be observed by the presence of Austroasiatic loanwords within Indo-Aryan languages.
Southeast Asians

A 2020 genetic study about Southeast Asian populations, found that mostly all Southeast Asians are closely related to East Asians and have mostly "East Asian-related" ancestry.
Ancient remains of hunter-gatherers in Maritime Southeast Asia, such as one Holocene hunter-gatherer from South Sulawesi, had ancestry from both, an Australasian lineage (represented by Papuans and Aboriginal Australasians) and an "Ancient Asian" lineage (represented by East Asians or Andamanese Onge). The hunter-gatherer individual had approximately c. 50% "Basal-East Asian" ancestry and c. 50% Australasian/Papuan ancestry, and was positioned in between modern East Asians and Papuans of Oceania. The authors concluded that East Asian-related ancestry expanded from Mainland Southeast Asia into Maritime Southeast Asia much earlier than previously suggested, as early as 25,000 BCE, long before the expansion of Austroasiatic and Austronesian groups.
A 2022 genetic study confirmed the close link between East Asians and Southeast Asians, which the authors term "East/Southeast Asian" (ESEA) populations, and also found a low but consistent proportion of South Asian-associated "SAS ancestry" (best samplified by modern Bengalis from Dhaka, Bangladesh) among specific Mainland Southeast Asian (MESA) ethnic groups (~2–16% as inferred by qpAdm), likely as a result of cultural diffision; mainly of South Asian merchants spreading Hinduism and Buddhism among the Indianized kingdoms of Southeast Asia. The authors however caution that Bengali samples harbor detectable East Asian ancestry, which may affect the estimation of shared haplotypes. Overall, the geneflow event is estimated to have happened between 500 and 1000 YBP.

Australasians
Main articles: Indigenous people of New Guinea, Melanesians, and Aboriginal AustraliansMelanesians and Aboriginal Australians are deeply related to East Asians. Genetic studies have revealed that Australasians descended from the same Eastern Eurasian source population as East Asians and indigenous South Asians (AASI).
References
- ^ Bennett, Andrew E.; Liu, Yichen; Fu, Qiaomei (4 December 2024). "Reconstructing the Human Population History of East Asia through Ancient Genomics". Elements in Ancient East Asia. doi:10.1017/9781009246675. ISBN 978-1-009-24667-5.
- Wang CC, Li H (June 2013). "Inferring human history in East Asia from Y chromosomes". Investigative Genetics. 4 (1): 11. doi:10.1186/2041-2223-4-11. PMC 3687582. PMID 23731529.
- Di D, Sanchez-Mazas A, Currat M (November 2015). "Computer simulation of human leukocyte antigen genes supports two main routes of colonization by human populations in East Asia". BMC Evolutionary Biology. 15 (1): 240. Bibcode:2015BMCEE..15..240D. doi:10.1186/s12862-015-0512-0. PMC 4632674. PMID 26530905.
- Vallini L, Marciani G, Aneli S, Bortolini E, Benazzi S, Pievani T, Pagani L (April 2022). "Genetics and Material Culture Support Repeated Expansions into Paleolithic Eurasia from a Population Hub Out of Africa". Genome Biology and Evolution. 14 (4). doi:10.1093/gbe/evac045. PMC 9021735. PMID 35445261.
- Seguin-Orlando, Andaine; Korneliussen, Thorfinn S.; Sikora, Martin; et al. (2014). "Genomic structure in Europeans dating back at least 36,200 years". Science. 346 (6213): 1113–1118. Bibcode:2014Sci...346.1113S. doi:10.1126/science.aaa0114. PMID 25378462 – via Science.org.
- Vallini, Leonardo; Zampieri, Carlo; Shoaee, Mohamed Javad; Bortolini, Eugenio; Marciani, Giulia; Aneli, Serena; Pievani, Telmo; Benazzi, Stefano; Barausse, Alberto; Mezzavilla, Massimo; Petraglia, Michael D.; Pagani, Luca (25 March 2024). "The Persian plateau served as hub for Homo sapiens after the main out of Africa dispersal". Nature Communications. 15 (1): 1882. Bibcode:2024NatCo..15.1882V. doi:10.1038/s41467-024-46161-7. ISSN 2041-1723. PMC 10963722. PMID 38528002.
- ^ Yang MA (6 January 2022). "A genetic history of migration, diversification, and admixture in Asia". Human Population Genetics and Genomics. 2 (1): 1–32. doi:10.47248/hpgg2202010001. ISSN 2770-5005.
...In contrast, mainland East and Southeast Asians and other Pacific islanders (e.g., Austronesian speakers) are closely related to each other and here denoted as belonging to an East and Southeast Asian (ESEA) lineage (Box 2). …the ESEA lineage differentiated into at least three distinct ancestries: Tianyuan ancestry which can be found 40,000–33,000 years ago in northern East Asia, ancestry found today across present-day populations of East Asia, Southeast Asia, and Siberia, but whose origins are unknown, and Hòabìnhian ancestry found 8,000–4,000 years ago in Southeast Asia, but whose origins in the Upper Paleolithic are unknown.
- Genetics and material culture support repeated expansions into Paleolithic Eurasia from a population hub out of Africa, Vallini et al. 2022 (4 April 2022) Quote: "Taken together with a lower bound of the final settlement of Sahul at 37 ka (the date of the deepest population splits estimated by Malaspinas et al. 2016), it is reasonable to describe Papuans as either an almost even mixture between East Asians and a lineage basal to West and East Asians occurred sometimes between 45 and 38 ka, or as a sister lineage of East Asians with or without a minor basal OoA or xOoA contribution. We here chose to parsimoniously describe Papuans as a simple sister group of Tianyuan, cautioning that this may be just one out of six equifinal possibilities."
- "Almost all living people outside of Africa trace back to a single migration more than 50,000 years ago". science.org. Retrieved 19 August 2022.
- Metspalu M, Kivisild T, Metspalu E, Parik J, Hudjashov G, Kaldma K, et al. (August 2004). "Most of the extant mtDNA boundaries in south and southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans". BMC Genetics. 5 (1): 26. doi:10.1186/1471-2156-5-26. PMC 516768. PMID 15339343.
- ^ Osada N, Kawai Y (2021). "Exploring models of human migration to the Japanese archipelago using genome-wide genetic data". Anthropological Science. 129 (1): 45–58. doi:10.1537/ase.201215. S2CID 234247309.
Via the southern route, ancestors of current Asian populations reached Southeast Asia and a part of Oceania around 70000–50000 years ago, probably through a coastal dispersal route (Bae et al., 2017). The oldest samples providing the genetic evidence of the northern migration route come from a high-coverage genome sequence of individuals excavated from the Yana RHS site in northeastern Siberia (Figure 2), which is about 31600 years old (Sikora et al., 2019). A wide range of artifacts, including bone crafts of wooly rhinoceros and mammoths, were excavated at the site (Pitulko et al., 2004). The analysis of genome sequences showed that the samples were deeply diverged from most present-day East Asians and more closely related to present-day Europeans, suggesting that the population reached the area through a route different from the southern route. A 24000-year-old individual excavated near Lake Baikal (Figure 2), also known as the Mal'ta boy, and 17000-year-old individuals from the Afontova Gora II site (Afontova Gora 2 and 3) showed similar genetic features to the Yana individuals (Raghavan et al., 2014; Fu et al., 2016; Sikora et al., 2019). Interestingly, genetic data suggested that Yana individuals received a large amount of gene flow from the East Asian lineage (Sikora et al., 2019; Yang et al., 2020).
- Gakuhari T, Nakagome S, Rasmussen S, Allentoft ME, Sato T, Korneliussen T, et al. (August 2020). "Ancient Jomon genome sequence analysis sheds light on migration patterns of early East Asian populations". Communications Biology. 3 (1): 437. doi:10.1038/s42003-020-01162-2. PMC 7447786. PMID 32843717.
Population genomic studies on present-day humans7,8 have exclusively supported the southern route origin of East Asian populations.
- Larena M, Sanchez-Quinto F, Sjödin P, McKenna J, Ebeo C, Reyes R, et al. (March 2021). "Multiple migrations to the Philippines during the last 50,000 years". Proceedings of the National Academy of Sciences of the United States of America. 118 (13): e2026132118. Bibcode:2021PNAS..11826132L. doi:10.1073/pnas.2026132118. PMC 8020671. PMID 33753512.
- Bae CJ, Douka K, Petraglia MD (December 2017). "On the origin of modern humans: Asian perspectives". Science. 358 (6368): eaai9067. doi:10.1126/science.aai9067. PMID 29217544. S2CID 4436271.
- Aoki K, Takahata N, Oota H, Wakano JY, Feldman MW (August 2023). "Infectious diseases may have arrested the southward advance of microblades in Upper Palaeolithic East Asia". Proceedings. Biological Sciences. 290 (2005): 20231262. doi:10.1098/rspb.2023.1262. PMC 10465978. PMID 37644833.
A single major migration of modern humans into the continents of Asia and Sahul was strongly supported by earlier studies using mitochondrial DNA, the non-recombining portion of Y chromosomes, and autosomal SNP data . Ancestral Ancient South Indians with no West Eurasian relatedness, East Asians, Onge (Andamanese hunter–gatherers) and Papuans all derive in a short evolutionary time from the eastward dispersal of an out-of-Africa population , although Europeans and East Asians are suggested to share more recent common ancestors than with Papuans . The HUGO (Human Genome Organization) Pan-Asian SNP consortium investigated haplotype diversity within present-day Asian populations and found a strong correlation with latitude, with diversity decreasing from south to north. The correlation continues to hold when only mainland Southeast Asian and East Asian populations are considered, and is perhaps attributable to a serial founder effect . These observations are consistent with the view that soon after the single eastward migration of modern humans, East Asians diverged in southern East Asia and dispersed northward across the continent.
- Demeter, Fabrice; Shackelford, Laura L.; Bacon, Anne-Marie; Duringer, Philippe; Westaway, Kira; Sayavongkhamdy, Thongsa; Braga, José; Sichanthongtip, Phonephanh; Khamdalavong, Phimmasaeng; Ponche, Jean-Luc; Wang, Hong; Lundstrom, Craig; Patole-Edoumba, Elise; Karpoff, Anne-Marie (4 September 2012). "Anatomically modern human in Southeast Asia (Laos) by 46 ka". Proceedings of the National Academy of Sciences. 109 (36): 14375–14380. Bibcode:2012PNAS..10914375D. doi:10.1073/pnas.1208104109. ISSN 0027-8424. PMC 3437904. PMID 22908291.
Inferences from nuclear (51), Y chromosome (52), and mitochondrial genome (53) data support an early migration of modern humans out of Africa and into Southeast Asia using a southern route by at least 60 ka. Patterns of genetic variation in recent human populations (11, 54, 55) recognize Southeast Asia as an important source for the peopling of East Asia and Australasia via a rapid, early settlement.
- Xu D, Li H (2017). Languages and Genes in Northwestern China and Adjacent Regions. Springer. p. 27. ISBN 978-981-10-4169-3. "In the study of Zhong et al. haplogroups O-M175, C-M130, D-M174 and N-M231 still suggests the substantial contribution of the southern route. However, the Central Asia and West Eurasia related haplogroups, such as haplogroups R-M207 and Q-M242, occur primarily in northwestern East Asia and their frequencies gradually decrease from west to east. In addition, the Y-STR diversities of haplogroups R-M207 and Q-M242 also indicate the existence of northern route migration about 18,000 years ago from Central Asia to North Asia, and recent population admixture along the Silk Road since about 3000 years ago (Piazza 1998)."
- Zhang X, Ji X, Li C, Yang T, Huang J, Zhao Y, et al. (July 2022). "A Late Pleistocene human genome from Southwest China". Current Biology. 32 (14): 3095–3109.e5. Bibcode:2022CBio...32E3095Z. doi:10.1016/j.cub.2022.06.016. PMID 35839766. S2CID 250502011. "In addition to the earliest southern settlement of AMHs in East Asia, ancient migration (40–18 kya) into East Asia via the “Northern Route” from West Eurasia was previously proposed. The “Northern Route” hypothesis would also explain where the subtle shared ancient north Eurasian (ANE) ancestry came from that is then also shared with Native Americans."
- Sato T, Adachi N, Kimura R, Hosomichi K, Yoneda M, Oota H, et al. (September 2021). "Whole-Genome Sequencing of a 900-Year-Old Human Skeleton Supports Two Past Migration Events from the Russian Far East to Northern Japan". Genome Biology and Evolution. 13 (9): evab192. doi:10.1093/gbe/evab192. PMC 8449830. PMID 34410389.
- ^ Huang, Xiufeng; Xia, Zi-Yang; Bin, Xiaoyun; He, Guanglin; Guo, Jianxin; Lin, Chaowen; Yin, Lianfei; Zhao, Jing; Ma, Zhuofei (8 November 2020), Genomic Insights into the Demographic History of Southern Chinese, doi:10.1101/2020.11.08.373225
- ^ Zhang, Ming; Fu, Qiaomei (1 June 2020). "Human evolutionary history in Eastern Eurasia using insights from ancient DNA". Current Opinion in Genetics & Development. Genetics of Human Origin. 62: 78–84. doi:10.1016/j.gde.2020.06.009. ISSN 0959-437X. PMID 32688244. S2CID 220671047.
- Watanabe Y, Ohashi J (8 March 2021). "Comprehensive analysis of Japanese archipelago population history by detecting ancestry-marker polymorphisms without using ancient DNA data". bioRxiv 10.1101/2020.12.07.414037.
- de Boer E (2020). "Japan considered from the hypothesis of farmer/language spread". Evolutionary Human Sciences. 2: e13. doi:10.1017/ehs.2020.7. PMC 10427481. PMID 37588377. S2CID 218926428.
- Downes, Natasha (21 January 2019). "Genetic study provides novel insights into the evolution of skin colour". UCL News (Press release). University College London. Retrieved 4 December 2021.
- Huang, Xin (2021). "Dissecting dynamics and differences of selective pressures in the evolution of human pigmentation". Biology Open. 10 (2). doi:10.1242/bio.056523. PMC 7888712. PMID 33495209.
- He, Guang-Lin; Wang, Meng-Ge; Zou, Xing; Yeh, Hui-Yuan; Liu, Chang-Hui; Liu, Chao; Chen, Gang; Wang, Chuan-Chao (2023). "Extensive ethnolinguistic diversity at the crossroads of North China and South Siberia reflects multiple sources of genetic diversity". Journal of Systematics and Evolution. 61 (1): 230–250. doi:10.1111/jse.12827. ISSN 1674-4918.
- Mao X, Zhang H, Qiao S, Liu Y, Chang F, Xie P, et al. (June 2021). "The deep population history of northern East Asia from the Late Pleistocene to the Holocene". Cell. 184 (12): 3256–3266.e13. doi:10.1016/j.cell.2021.04.040. PMID 34048699. S2CID 235226413.
- Qin, Pengfei; Zhou, Ying; Lou, Haiyi; Lu, Dongsheng; Yang, Xiong; Wang, Yuchen; Jin, Li; Chung, Yeun-Jun; Xu, Shuhua (2 April 2015). "Quantitating and Dating Recent Gene Flow between European and East Asian Populations". Scientific Reports. 5 (1): 9500. Bibcode:2015NatSR...5.9500Q. doi:10.1038/srep09500. ISSN 2045-2322. PMC 4382708. PMID 25833680.
- ^ Tagore D, Aghakhanian F, Naidu R, Phipps ME, Basu A (March 2021). "Insights into the demographic history of Asia from common ancestry and admixture in the genomic landscape of present-day Austroasiatic speakers". BMC Biology. 19 (1): 61. doi:10.1186/s12915-021-00981-x. PMC 8008685. PMID 33781248.
- "Direct genetic evidence of founding population reveals story of first Native Americans". University of Cambridge. 3 January 2018. Retrieved 9 June 2021.
- Liu D, Duong NT, Ton ND, Van Phong N, Pakendorf B, Van Hai N, Stoneking M (September 2020). "Extensive Ethnolinguistic Diversity in Vietnam Reflects Multiple Sources of Genetic Diversity". Molecular Biology and Evolution. 37 (9): 2503–2519. doi:10.1093/molbev/msaa099. PMC 7475039. PMID 32344428.
- Liu, Dang; Duong, Nguyen Thuy; Ton, Nguyen Dang; Phong, Nguyen Van; Pakendorf, Brigitte; Hai, Nong Van; Stoneking, Mark (28 November 2019), Extensive ethnolinguistic diversity in Vietnam reflects multiple sources of genetic diversity, doi:10.1101/857367, hdl:21.11116/0000-0006-4AD8-4, retrieved 14 November 2024
- Wong EH, Khrunin A, Nichols L, Pushkarev D, Khokhrin D, Verbenko D, et al. (January 2017). "Reconstructing genetic history of Siberian and Northeastern European populations". Genome Research. 27 (1): 1–14. doi:10.1101/gr.202945.115. PMC 5204334. PMID 27965293.
- Sikora, Martin; Pitulko, Vladimir V.; Sousa, Vitor C.; Allentoft, Morten E.; Vinner, Lasse; Rasmussen, Simon; Margaryan, Ashot; Damgaard, Peter de Barros; Castro, Constanza de la Fuente (22 October 2018), The population history of northeastern Siberia since the Pleistocene, doi:10.1101/448829, hdl:1887/3198847, retrieved 14 November 2024
- "Xiongnu People". britannica.com. Encyclopædia Britannica. Archived from the original on 11 March 2020. Retrieved 25 July 2015.
- di Cosmo 2004: 186
- ^ Savelyev, Alexander; Jeong, Choongwoon (7 May 2020). "Early nomads of the Eastern Steppe and their tentative connections in the West". Evolutionary Human Sciences. 2 (E20). doi:10.1017/ehs.2020.18. hdl:21.11116/0000-0007-772B-4. PMC 7612788. PMID 35663512. S2CID 218935871.
 Text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. "Such a distribution of Xiongnu words may be an indication that both Turkic and Eastern Iranian-speaking groups were present among the Xiongnu in the earlier period of their history. Etymological analysis shows that some crucial components in the Xiongnu political, economic and cultural package, including dairy pastoralism and elements of state organization, may have been imported by the Eastern Iranians. Arguably, these Iranian-speaking groups were assimilated over time by the predominant Turkic-speaking part of the Xiongnu population. ... The genetic profile of published Xiongnu individuals speaks against the Yeniseian hypothesis, assuming that modern Yeniseian speakers (i.e. Kets) are representative of the ancestry components in the historical Yeniseian speaking groups in southern Siberia. In contrast to the Iron Age populations listed in Table 2, Kets do not have the Iranian-related ancestry component but harbour a strong genetic affinity with Samoyedic-speaking neighbours, such as Selkups (Jeong et al., 2018, 2019)."
Text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. "Such a distribution of Xiongnu words may be an indication that both Turkic and Eastern Iranian-speaking groups were present among the Xiongnu in the earlier period of their history. Etymological analysis shows that some crucial components in the Xiongnu political, economic and cultural package, including dairy pastoralism and elements of state organization, may have been imported by the Eastern Iranians. Arguably, these Iranian-speaking groups were assimilated over time by the predominant Turkic-speaking part of the Xiongnu population. ... The genetic profile of published Xiongnu individuals speaks against the Yeniseian hypothesis, assuming that modern Yeniseian speakers (i.e. Kets) are representative of the ancestry components in the historical Yeniseian speaking groups in southern Siberia. In contrast to the Iron Age populations listed in Table 2, Kets do not have the Iranian-related ancestry component but harbour a strong genetic affinity with Samoyedic-speaking neighbours, such as Selkups (Jeong et al., 2018, 2019)."
- Savelyev & Jeong 2020:"Specifically, individuals from Iron Age steppe and Xiongnu have an ancestry related to present-day and ancient Iranian/Caucasus/Turan populations in addition to the ancestry components derived from the Late Bronze Age populations. We estimate that they derive between 5 and 25% of their ancestry from this new source, with 18% for Xiongnu (Table 2). We speculate that the introduction of this new western Eurasian ancestry may be linked to the Iranian elements in the Xiongnu linguistic material, while the Turkic-related component may be brought by their eastern Eurasian genetic substratum." Table 2: Sintashta_MLBA, 0.239; Khovsgol LBA, 0.582; Gonur1 BA 0.178
- ^ Damgaard PB, Marchi N, Rasmussen S, Peyrot M, Renaud G, Korneliussen T, et al. (May 2018). "137 ancient human genomes from across the Eurasian steppes". Nature. 557 (7705): 369–374. Bibcode:2018Natur.557..369D. doi:10.1038/s41586-018-0094-2. hdl:1887/3202709. PMID 29743675. S2CID 13670282.
- Lee, Juhyeon; Miller, Bryan K.; Bayarsaikhan, Jamsranjav; Johannesson, Erik; Miller, Alicia Ventresca; Warinner, Christina; Jeong, Choongwon (April 2023). "Genetic population structure of the Xiongnu Empire at imperial and local scales". Science Advances. 9 (15): eadf3904. Bibcode:2023SciA....9F3904L. doi:10.1126/sciadv.adf3904. PMC 10104459. PMID 37058560.
- Rogers & Kaestle 2022.
- Li H (2012). Y-Chromosome Genetic Diversity of the Ancient North Chinese populations (Report). China: Jilin University.
- Lee J, Miller BK, Bayarsaikhan J, Johannesson E, Ventresca Miller A, Warinner C, Jeong C (April 2023). "Genetic population structure of the Xiongnu Empire at imperial and local scales". Science Advances. 9 (15). American Association for the Advancement of Science (AAAS): eadf3904. Bibcode:2023SciA....9F3904L. doi:10.1126/sciadv.adf3904. PMC 10104459. PMID 37058560.
- Lee 2023: Haplogroup information for DA39 is located in Supplementary Materials document Data S1 (abk1900_Data_S1.xlsx), row 58. "Before this study, only one other individual from an elite square tomb had been analyzed in a genome-wide manner: DA39 from Tomb 1 at the imperial elite site of Gol Mod 2 in central-north Mongolia (13). This adult male, buried in one of the largest square tomb complexes excavated to date, surrounded by at least 27 satellite burials, and containing rare exotic items such as Roman glass bowls, was likely a chanyu, or ruler of the empire (73, 74)."
- Rogers LL, Kaestle FA (2022). "Analysis of mitochondrial DNA haplogroup frequencies in the population of the slab burial mortuary culture of Mongolia (ca. 1100–300 BCE )". American Journal of Biological Anthropology. 177 (4): 644–657. doi:10.1002/ajpa.24478. ISSN 2692-7691. S2CID 246508594. " The first pattern is that the slab burial mtDNA frequencies are extremely similar to those of the aggregated Xiongnu populations and relatively similar to those of the various Bronze Age Mongolian populations, strongly supporting a population continuity hypothesis for the region over these time periods (Honeychurch, 2013)"
- Keyser-Tracqui C, Crubézy E, Ludes B (August 2003). "Nuclear and mitochondrial DNA analysis of a 2,000-year-old necropolis in the Egyin Gol Valley of Mongolia". American Journal of Human Genetics. 73 (2): 247–60. doi:10.1086/377005. PMC 1180365. PMID 12858290.
- Lee JY, Kuang S (2017). "A Comparative Analysis of Chinese Historical Sources and Y-DNA Studies with Regard to the Early and Medieval Turkic Peoples". Inner Asia. 19 (2): 197–239. doi:10.1163/22105018-12340089. ISSN 1464-8172. S2CID 165623743. "Analysis of the mitochondrial DNA, which is maternally inherited, shows that the Xiongnu remains from this Egyin Gol necropolis consist mainly of Asian lineages (89%). West Eurasian lineages makeup the rest (11%) (Keyser-Tracqui et al. (2003: 258). However, according to a more recent study of ancient human remains from central Mongolia, the Xiongnu population in central Mongolia possessed a higher frequency of western mitochondrial DNA haplotypes (37.5%) than the Xiongnu from the Egyin Gol necropolis (Rogers 2016: 78)."
- Cai, Dawei; Zheng, Ying; Bao, Qingchuan; Hu, Xiaonong; Chen, Wenhu; Zhang, Fan; Cao, Jianen; Ning, Chao (24 November 2023). "Ancient DNA sheds light on the origin and migration patterns of the Xianbei confederation". Archaeological and Anthropological Sciences. 15 (12): 194. Bibcode:2023ArAnS..15..194C. doi:10.1007/s12520-023-01899-x. ISSN 1866-9565. S2CID 265381985.
- Li et al. 2018, p. 1.
- Changchun Y, Li X, Xiaolei Z, Hui Z, Hong Z (November 2006). "Genetic analysis on Tuoba Xianbei remains excavated from Qilang Mountain Cemetery in Qahar Right Wing Middle Banner of Inner Mongolia". FEBS Letters. 580 (26): 6242–6246. Bibcode:2006FEBSL.580.6242C. doi:10.1016/j.febslet.2006.10.030. PMID 17070809. S2CID 19492267.
- Wang H, Ge B, Mair VH, Cai D, Xie C, Zhang Q, et al. (November 2007). "Molecular genetic analysis of remains from Lamadong cemetery, Liaoning, China". American Journal of Physical Anthropology. 134 (3): 404–411. doi:10.1002/ajpa.20685. PMID 17632796.
- Zhou H (March 2014). "Genetic analyses of Xianbei populations about 1,500–1,800 years old". Human Genetics. 50 (3): 308–314. doi:10.1134/S1022795414030119. S2CID 18809679.
- Osada, Naoki; Kawai, Yosuke (2021). "Exploring models of human migration to the Japanese archipelago using genome-wide genetic data". Anthropological Science. 129 (1): 45–58. doi:10.1537/ase.201215.
- ^ Cooke, Niall P.; Mattiangeli, Valeria; Cassidy, Lara M.; Okazaki, Kenji; Stokes, Caroline A.; Onbe, Shin; Hatakeyama, Satoshi; Machida, Kenichi; Kasai, Kenji; Tomioka, Naoto; Matsumoto, Akihiko (September 2021). "Ancient genomics reveals tripartite origins of Japanese populations". Science Advances. 7 (38): eabh2419. Bibcode:2021SciA....7.2419C. doi:10.1126/sciadv.abh2419. PMC 8448447. PMID 34533991.
- McColl, Hugh; Racimo, Fernando; Vinner, Lasse; Demeter, Fabrice; Gakuhari, Takashi; et al. (2018). "The prehistoric peopling of Southeast Asia". Science. 361 (6397). American Association for the Advancement of Science (AAAS): 88–92. Bibcode:2018Sci...361...88M. doi:10.1126/science.aat3628. hdl:10072/383365. ISSN 0036-8075. PMID 29976827. S2CID 206667111.
- Yang, Melinda A. (6 January 2022). "A genetic history of migration, diversification, and admixture in Asia". Human Population Genetics and Genomics. 2 (1): 1–32. doi:10.47248/hpgg2202010001.
- 张明, 平婉菁; ZHANG Ming, PING Wanjing (15 June 2023). "古基因组揭示史前欧亚大陆现代人复杂遗传历史". 人类学学报 (in Chinese). 42 (3): 412. doi:10.16359/j.1000-3193/AAS.2023.0010. ISSN 1000-3193.
- ^ Gakuhari T, Nakagome S, Rasmussen S, Allentoft ME, Sato T, Korneliussen T, et al. (August 2020). "Ancient Jomon genome sequence analysis sheds light on migration patterns of early East Asian populations". Communications Biology. 3 (1): 437. doi:10.1038/s42003-020-01162-2. PMC 7447786. PMID 32843717.
- Zhang X, He G, Li W, Wang Y, Li X, Chen Y, et al. (30 September 2021). "Genomic Insight Into the Population Admixture History of Tungusic-Speaking Manchu People in Northeast China". Frontiers in Genetics. 12: 754492. doi:10.3389/fgene.2021.754492. PMC 8515022. PMID 34659368.
 This article incorporates text available under the CC BY 4.0 license.
This article incorporates text available under the CC BY 4.0 license.
- ^ Xue, Yali; Zerjal, Tatiana; Bao, Weidong; Zhu, Suling; Shu, Qunfang; Xu, Jiujin; Du, Ruofu; Fu, Songbin; Li, Pu; Hurles, Matthew E; Yang, Huanming; Tyler-Smith, Chris (1 April 2006). "Male Demography in East Asia: A North–South Contrast in Human Population Expansion Times". Genetics. 172 (4): 2431–2439. doi:10.1534/genetics.105.054270. PMC 1456369. PMID 16489223.
- ^ Wang Chi-zao, Shi Mei-sen, and Li Hui, "The Origin of Daur from the Perspective of Molecular Anthropology." Journal of North Minzu University 2018, No.5, Gen.No.143.
- Wei LH, Yan S, Yu G, Huang YZ, Yao DL, Li SL, et al. (March 2017). "Genetic trail for the early migrations of Aisin Gioro, the imperial house of the Qing dynasty". Journal of Human Genetics. 62 (3): 407–411. doi:10.1038/jhg.2016.142. PMID 27853133. S2CID 7685248.
- Yan S, Tachibana H, Wei LH, Yu G, Wen SQ, Wang CC (June 2015). "Y chromosome of Aisin Gioro, the imperial house of the Qing dynasty". Journal of Human Genetics. 60 (6): 295–8. arXiv:1412.6274. doi:10.1038/jhg.2015.28. PMID 25833470. S2CID 7505563.
- ^ "Did you know DNA was used to uncover the origin of the House of Aisin Gioro?". Did You Know DNA... 14 November 2016. Retrieved 5 November 2020.
- Xue Y, Zerjal T, Bao W, Zhu S, Lim SK, Shu Q, et al. (December 2005). "Recent spread of a Y-chromosomal lineage in northern China and Mongolia". American Journal of Human Genetics. 77 (6): 1112–6. doi:10.1086/498583. PMC 1285168. PMID 16380921.
- "Asian Ancestry based on Studies of Y-DNA Variation: Part 3. Recent demographics and ancestry of the male East Asians – Empires and Dynasties". Genebase Tutorials. Archived from the original on 25 November 2013.
- Zhang, Xianpeng; He, Guanglin; Li, Wenhui; Wang, Yunfeng; Li, Xin; Chen, Ying; Qu, Quanying; Wang, Ying; Xi, Huanjiu; Wang, Chuan-Chao; Wen, Youfeng (30 September 2021). "Genomic Insight Into the Population Admixture History of Tungusic-Speaking Manchu People in Northeast China". Frontiers in Genetics. 12: 754492. doi:10.3389/fgene.2021.754492. ISSN 1664-8021. PMC 8515022. PMID 34659368.
- Hammer, Michael F.; Karafet, Tatiana M.; Park, Hwayong; Omoto, Keiichi; Harihara, Shinji; Stoneking, Mark; Horai, Satoshi (January 2006). "Dual origins of the Japanese: common ground for hunter-gatherer and farmer Y chromosomes". Journal of Human Genetics. 51 (1): 47–58. doi:10.1007/s10038-005-0322-0. PMID 16328082. S2CID 6559289.
- "公益財団法人 アイヌ民族文化財団". www.ff-ainu.or.jp (in Japanese). Retrieved 8 December 2023.
- Jinam, Timothy; Nishida, Nao; Hirai, Momoki; Kawamura, Shoji; Oota, Hiroki; Umetsu, Kazuo; Kimura, Ryosuke; Ohashi, Jun; Tajima, Atsushi (December 2012). "The history of human populations in the Japanese Archipelago inferred from genome-wide SNP data with a special reference to the Ainu and the Ryukyuan populations". Journal of Human Genetics. 57 (12): 787–795. doi:10.1038/jhg.2012.114. PMID 23135232.
- "記者会見「日本列島3人類集団の遺伝的近縁性」". 東京大学 (in Japanese). Retrieved 8 November 2021.
- Mitsuru Sakitani (2009). 『DNA・考古・言語の学際研究が示す新・日本列島史』 [New History of the Japanese Islands Shown by Interdisciplinary Studies on DNA, Archeology, and Language] (in Japanese). Bensei Publishing. ISBN 9784585053941.
- ^ Suzuki, Yuka (6 December 2012). "Ryukyuan, Ainu People Genetically Similar". Asian Scientist. Archived from the original on 16 June 2016. Retrieved 11 June 2016.
- Cooke NP, Mattiangeli V, Cassidy LM, Okazaki K, Stokes CA, Onbe S, et al. (September 2021). "Ancient genomics reveals tripartite origins of Japanese populations". Science Advances. 7 (38): eabh2419. Bibcode:2021SciA....7.2419C. doi:10.1126/sciadv.abh2419. PMC 8448447. PMID 34533991.
- "'Jomon woman' helps solve Japan's genetic mystery | NHK WORLD-JAPAN News". NHK WORLD. Retrieved 19 November 2020.
- Jinam, Timothy; Nishida, Nao; Hirai, Momoki; Kawamura, Shoji; Oota, Hiroki; Umetsu, Kazuo; Kimura, Ryosuke; Ohashi, Jun; Tajima, Atsushi; Yamamoto, Toshimichi; Tanabe, Hideyuki; Mano, Shuhei; Suto, Yumiko; Kaname, Tadashi; Naritomi, Kenji (8 November 2012). "The history of human populations in the Japanese Archipelago inferred from genome-wide SNP data with a special reference to the Ainu and the Ryukyuan populations". Journal of Human Genetics. 57 (12): 787–795. doi:10.1038/jhg.2012.114. ISSN 1435-232X.
- 弥生人DNAで迫る日本人の起源」 [The origin of Japanese people approaching with Yayoi DNA]. ja:サイエンスZERO (Television production) (in Japanese). NHK. 23 December 2018.
- Horai, Satoshi; Murayama, Kumiko (1996). "mtDNA Polymorphism in East Asian Populations, with Special Reference to the Peopling of Japan". American Journal of Human Genetics. 59 (3). Cambridge, Massachusetts: Cell Press: 579–590. PMC 1914908. PMID 8751859.
- Boer, Elisabeth de; Yang, Melinda A.; Kawagoe, Aileen; Barnes, Gina L. (2020). "Japan considered from the hypothesis of farmer/language spread". Evolutionary Human Sciences. 2: e13. doi:10.1017/ehs.2020.7. ISSN 2513-843X. PMC 10427481. PMID 37588377.
- ^ Wang, Chuan-Chao (2021). "Genomic insights into the formation of human populations in East Asia". Nature. 591 (7850): 413–419. Bibcode:2021Natur.591..413W. doi:10.1038/s41586-021-03336-2. PMC 7993749. PMID 33618348.
- Nikkei Science (23 June 2021). "渡来人、四国に多かった? ゲノムが明かす日本人ルーツ" [Were there many migrants in Shikoku? Japanese roots revealed by genome analysis]. nikkei.com (in Japanese). Retrieved 1 May 2022.
- Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, et al. (November 2000). "Y chromosome sequence variation and the history of human populations". Nature Genetics. 26 (3): 358–61. doi:10.1038/81685. PMID 11062480. S2CID 12893406.
- The JPT sample is considered "as generally representative of the majority population in Japan". See Matsuda I. "Japanese in Tokyo, Japan – Population Description". Camden, NJ: Coriell Institute for Medical Research.
- Poznik GD, Xue Y, Mendez FL, Willems TF, Massaia A, Wilson Sayres MA, et al. (June 2016). "Punctuated bursts in human male demography inferred from 1,244 worldwide Y-chromosome sequences". Nature Genetics. 48 (6): 593–9. doi:10.1038/ng.3559. hdl:11858/00-001M-0000-002A-F024-C. PMC 4884158. PMID 27111036.
- Zheng HX, Yan S, Qin ZD, Wang Y, Tan JZ, Li H, Jin L (2011). "Major population expansion of East Asians began before neolithic time: evidence of mtDNA genomes". PLOS ONE. 6 (10): e25835. Bibcode:2011PLoSO...625835Z. doi:10.1371/journal.pone.0025835. PMC 3188578. PMID 21998705.
- Preucel, Robert; Mrozowski, Stephen; Nelson, Sarah (2010). Contemporary Archaeology in Theory: The New Pragmatism (2nd ed.). Wiley-Blackwell. pp. 218–221.
- 水野, 文月 (15 October 2024). "弥生時代人の古代ゲノム解析から渡来人のルーツを探る". 東京大学 大学院理学系研究科・理学部 (in Japanese).
- ^ Wang, Rui; Wang, Chuan-Chao (8 August 2022). "Human genetics: The dual origin of Three Kingdoms period Koreans". Current Biology. 32 (15): R844 – R847. Bibcode:2022CBio...32.R844W. doi:10.1016/j.cub.2022.06.044. ISSN 0960-9822. PMID 35944486. S2CID 251410856.
- Siska, Veronika; Jones, Eppie; Jeon, Sungwon; et al. (2017). "Genome-wide data from two early Neolithic East Asian individuals dating to 7700 years ago". Science Advances. 3 (2): e1601877. Bibcode:2017SciA....3E1877S. doi:10.1126/sciadv.1601877. hdl:2262/90843. PMC 5287702. PMID 28164156.
- Jin HJ, Tyler-Smith C, Kim W (16 January 2009). "The peopling of Korea revealed by analyses of mitochondrial DNA and Y-chromosomal markers". PLOS ONE. 4 (1): e4210. Bibcode:2009PLoSO...4.4210J. doi:10.1371/journal.pone.0004210. PMC 2615218. PMID 19148289.
- ^ Kim SH, Kim KC, Shin DJ, Jin HJ, Kwak KD, Han MS, Song JM, Kim W, Kim W (April 2011). "High frequencies of Y-chromosome haplogroup O2b-SRY465 lineages in Korea: a genetic perspective on the peopling of Korea". Investigative Genetics. 2 (1): 10. doi:10.1186/2041-2223-2-10. PMC 3087676. PMID 21463511.
 Text was copied from this source, which is available under a Creative Commons Attribution 2.0 (CC BY 2.0) license.
Text was copied from this source, which is available under a Creative Commons Attribution 2.0 (CC BY 2.0) license.
- Wang & Wang 2022: "The genetic legacy of Jomon was not restricted to Japan but was also found in Neolithic Korea."
- Wang & Wang 2022: "Taking no account of this Bronze Age sample, the possible scenario was that the indigenous Jomon-related ancestry was largely replaced through admixture with the incoming northern China populations and disappeared in the later centuries, creating relatively homogeneous present-day Koreans."
- Wang & Wang 2022: "Present-day Koreans are a highly homogeneous population without apparent genetic substructure on a whole-genome scale and with close genetic relationships with Japanese and northern Han Chinese. The genetic variation of the Y chromosome and mitochondrial DNA show that Koreans contain lineages typical of both Southeast and Northeast Asian populations, suggesting that Korea was populated in multiple waves"
- Lee, Don-Nyeong; Jeon, Chae Lin; Kang, Jiwon; Burri, Marta; Krause, Johannes; Woo, Eun Jin; Jeong, Choongwon (December 2022). "Genomic detection of a secondary family burial in a single jar coffin in early Medieval Korea". American Journal of Biological Anthropology. 179 (4): 585–597. doi:10.1002/ajpa.24650. ISSN 2692-7691. PMC 9827920. "In both ancient and present-day Koreans, we do not detect a statistically significant contribution from the Jomon hunter-gatherer gene pool of the Japanese archipelago (Table S7A), although previous studies report occasional presence of the Jomon ancestry contribution from Neolithic to the early Medieval period (Gelabert et al., 2022; Robbeets et al., 2021). When we replace the genetic northern proxy from WLR_BA to Middle Neolithic individuals from the Miaogizou site in Inner Mongolia (“Miaozigou_MN”), we detect a small but significant amount of Jomon contribution in the Gunsan individuals and present-day Ulsan Koreans (3.1%–4.4%; Table S7B). We believe that WLR_BA provides a more suitable model for ancient and present-day Koreans given its geographical and temporal proximity to them. The remaining well-fitting source pairs provide qualitatively similar results (Table S8)."
- Kim, Jungeun; Jeon, Sungwon; Choi, Jae-Pil; et al. (2020). "The Origin and Composition of Korean Ethnicity Analyzed by Ancient and Present-Day Genome Sequences". Genome Biology and Evolution. 12 (5): 553–565. doi:10.1093/gbe/evaa062. PMC 7250502. PMID 32219389.
- Kim SH, Han MS, Kim W, Kim W (November 2010). "Y chromosome homogeneity in the Korean population". International Journal of Legal Medicine. 124 (6): 653–7. doi:10.1007/s00414-010-0501-1. PMID 20714743. S2CID 27125545.
- ^ So Yeun Kwon, Hwan Young Lee, Eun Young Lee, Woo Ick Yang, and Kyoung-Jin Shin, "Confirmation of Y haplogroup tree topologies with newly suggested Y-SNPs for the C2, O2b and O3a subhaplogroups." Forensic Science International: Genetics 19 (2015) 42–46. http://dx.doi.org/10.1016/j.fsigen.2015.06.003
- Zhang, Ai Hua; Lee, Hye Young; Seo, Seung Bum; Lee, Hyo Jung; Jin, Hong Xuan; Cho, So Hee; Lyoo, Sung Hee; Kim, Ki Ha; Lee, Jae Won; Lee, Soong Deok (29 May 2012). "Y Haplogroup Distribution in Korean and Other Populations". Korean Journal of Legal Medicine. 36 (1): 34–44. doi:10.7580/KoreanJLegMed.2012.36.1.34.
- Choi, Sun Seong; Park, Kyung Hwa; Nam, Da Eun; Kang, Tae Hoon; Chung, Ki Wha (1 December 2017). "Y-chromosome haplogrouping for Asians using Y-SNP target sequencing". Forensic Science International: Genetics Supplement Series. 6: e235 – e237. doi:10.1016/j.fsigss.2017.09.100. ISSN 1875-1768.
- Kim, Soon Hee; Han, Myun Soo; Kim, Wook; Kim, Won (November 2010). "Y chromosome homogeneity in the Korean population". International Journal of Legal Medicine. 124 (6): 653–657. doi:10.1007/s00414-010-0501-1. ISSN 1437-1596. PMID 20714743. S2CID 27125545.
- Jin, Han-Jun; Tyler-Smith, Chris; Kim, Wook (16 January 2009). "The Peopling of Korea Revealed by Analyses of Mitochondrial DNA and Y-Chromosomal Markers". PLOS ONE. 4 (1): e4210. Bibcode:2009PLoSO...4.4210J. doi:10.1371/journal.pone.0004210. ISSN 1932-6203. PMC 2615218. PMID 19148289.
- Jeon, Sungwon; Bhak, Youngjune; Choi, Yeonsong; Jeon, Yeonsu; Kim, Seunghoon; Jang, Jaeyoung; Jang, Jinho; Blazyte, Asta; Kim, Changjae; Kim, Yeonkyung; Shim, Jungae; Kim, Nayeong; Kim, Yeo Jin; Park, Seung Gu; Kim, Jungeun (27 May 2020). "Korean Genome Project: 1094 Korean personal genomes with clinical information". Science Advances. 6 (22): eaaz7835. Bibcode:2020SciA....6.7835J. doi:10.1126/sciadv.aaz7835. ISSN 2375-2548. PMC 7385432. PMID 32766443.
- Hong, Seung Beom; Kim, Ki Cheol; Kim, Wook (1 October 2014). "Mitochondrial DNA haplogroups and homogeneity in the Korean population". Genes & Genomics. 36 (5): 583–590. doi:10.1007/s13258-014-0194-9. ISSN 2092-9293. S2CID 256067815.
- "Population dynamics and the rise of empires in Inner Asia: Genome-wide analysis spanning 6,000 years in the eastern Eurasian Steppe gives insights to the formation of Mongolia's empires". ScienceDaily. Retrieved 4 June 2023.
- Yang, Xiaomin; Sarengaowa; He, Guanglin; Guo, Jianxin; Zhu, Kongyang; Ma, Hao; Zhao, Jing; Yang, Meiqing; Chen, Jing; Zhang, Xianpeng; Tao, Le; Liu, Yilan; Zhang, Xiu-Fang; Wang, Chuan-Chao (2021). "Genomic Insights Into the Genetic Structure and Natural Selection of Mongolians". Frontiers in Genetics. 12: 735786. doi:10.3389/fgene.2021.735786. ISSN 1664-8021. PMC 8693022. PMID 34956310.
- "DNA Match Solves Ancient Mystery". china.org.cn. Retrieved 25 December 2018.
- Malyarchuk BA, Derenko M, Denisova G, Woźniak M, Rogalla U, Dambueva I, Grzybowski T (June 2016). "Y chromosome haplotype diversity in Mongolic-speaking populations and gene conversion at the duplicated STR DYS385a,b in haplogroup C3-M407". Journal of Human Genetics. 61 (6): 491–496. doi:10.1038/jhg.2016.14. PMID 26911356. S2CID 13217444.
- Guang‐Lin He, Meng‐Ge Wang, Xing Zou, Hui‐Yuan Yeh, Chang‐Hui Liu, Chao Liu, Gang Chen, and Chuan‐Chao Wang. "Extensive ethnolinguistic diversity at the crossroads of North China and South Siberia reflects multiple sources of genetic diversity." J Syst Evol, 2023, 61(1): 230-250. https://doi.org/10.1111/jse.12827
- "Mongol Genetics – DNA of Mongolia's Khalkha Mongolians and others". khazaria.com. Retrieved 25 December 2018.
- Kolman CJ, Sambuughin N, Bermingham E (April 1996). "Mitochondrial DNA analysis of Mongolian populations and implications for the origin of New World founders". Genetics. 142 (4): 1321–34. doi:10.1093/genetics/142.4.1321. PMC 1207128. PMID 8846908.
- Cheng B, Tang W, He L, Dong Y, Lu J, Lei Y, et al. (October 2008). "Genetic imprint of the Mongol: signal from phylogeographic analysis of mitochondrial DNA". Journal of Human Genetics. 53 (10): 905–913. doi:10.1007/s10038-008-0325-8. PMID 18769869.
European-prevalent haplogroups (HV, U, K, I, J) are 14.3% in Xinjiang Mongolian, 10% in Mongolia, 8.4% in central Inner Mongolian samples, and only 2% in eastern Xin Barage Zuoqi County samples, showing decreasing frequencies from west to east
- Feng, Qidi; Lu, Yan; Ni, X.; Yuan, K.; Yang, Ya-jun; Yang, Xiong; Liu, Chang; Lou, H.; Ning, Zhilin; Wang, Yuchen; Lu, Dongsheng; Zhang, Chao; Zhou, Ying; Shi, Meng; Tian, L. (2017). "Genetic History of Xinjiang's Uyghurs Suggests Bronze Age Multiple-Way Contacts in Eurasia". Molecular Biology and Evolution. 34 (10): 2572–2582. doi:10.1093/molbev/msx177. PMID 28595347. S2CID 28730957.
- ^ Wang, Yuchen; Lu Dongsheng; Chung Yeun-Jun; Xu Shuhua (2018). "Genetic structure, divergence and admixture of Han Chinese, Japanese and Korean populations". Hereditas. 155: 19. doi:10.1186/s41065-018-0057-5. PMC 5889524. PMID 29636655.
- ^ Yang, Melinda A.; Fan, Xuechun; Sun, Bo; Chen, Chungyu; Lang, Jianfeng; Ko, Ying-Chin; Tsang, Cheng-hwa; Chiu, Hunglin; Wang, Tianyi; Bao, Qingchuan; Wu, Xiaohong; Hajdinjak, Mateja; Ko, Albert Min-Shan; Ding, Manyu; Cao, Peng (17 July 2020). "Ancient DNA indicates human population shifts and admixture in northern and southern China". Science. 369 (6501): 282–288. Bibcode:2020Sci...369..282Y. doi:10.1126/science.aba0909. ISSN 0036-8075. PMID 32409524. S2CID 218649510.
- Cao, Yanan; Li, Lin; Xu, Min; et al. (2020). "The ChinaMAP analytics of deep whole genome sequences in 10,588 individuals". Cell Research. 30 (9): 717–731. doi:10.1038/s41422-020-0322-9. PMC 7609296. PMID 32355288.
- ^ Pan, Ziqing; Xu, Shuhua (2019). "Population genomics of East Asian ethnic groups". Hereditas. 157 (49). Berlin: BioMed Central (published 2020): 49. doi:10.1186/s41065-020-00162-w. PMC 7724877. PMID 33292737.
- Shi, Cheng-Min; Liu, Qi; Zhao, Shilei; Chen, Hua (21 March 2019). "Ancestry informative SNP panels for discriminating the major East Asian populations: Han Chinese, Japanese and Korean". Annals of Human Genetics. 29 (2). Cambridge: John Wiley & Sons: 348–354. doi:10.1111/ahg.12320. PMID 31025319. Archived from the original on 5 November 2021. Retrieved 14 September 2021.
- ^ Zhao, Yong-Bin; Zhang, Ye; Zhang, Quan-Chao; Li, Hong-Jie; Cui, Ying-Qiu; Xu, Zhi; Jin, Li; Zhou, Hui; Zhu, Hong (4 May 2015). Hofreiter, Michael (ed.). "Ancient DNA Reveals That the Genetic Structure of the Northern Han Chinese Was Shaped Prior to 3,000 Years Ago". PLOS ONE. 10 (5): e0125676. Bibcode:2015PLoSO..1025676Z. doi:10.1371/journal.pone.0125676. PMC 4418768. PMID 25938511.
- Ebrey, Patricia Buckley (2023). "Rethinking Han Chinese Identity". China Review. 23 (2): 57–86. ISSN 1680-2012. JSTOR 48726991.
- Feng, Qidi; Lu, Yan; Ni, X.; Yuan, K.; Yang, Ya-jun; Yang, Xiong; Liu, Chang; Lou, H.; Ning, Zhilin; Wang, Yuchen; Lu, Dongsheng; Zhang, Chao; Zhou, Ying; Shi, Meng; Tian, L. (2017). "Genetic History of Xinjiang's Uyghurs Suggests Bronze Age Multiple-Way Contacts in Eurasia". Molecular Biology and Evolution. 34 (10): 2572–2582. doi:10.1093/molbev/msx177. PMID 28595347. S2CID 28730957.
- Zhao, Yong-Bin; Zhang, Ye; Zhang, Quan-Chao; Li, Hong-Jie; Cui, Ying-Qiu; Xu, Zhi; Jin, Li; Zhou, Hui; Zhu, Hong (4 May 2015). "Ancient DNA Reveals That the Genetic Structure of the Northern Han Chinese Was Shaped Prior to 3,000 Years Ago". PLOS ONE. 10 (5): e0125676. Bibcode:2015PLoSO..1025676Z. doi:10.1371/journal.pone.0125676. PMC 4418768. PMID 25938511.
- Chiang, Charleston W.K.; Mangul, Serghei; Robles, Christopher; Sankararaman, Sriram (2018). "A Comprehensive Map of Genetic Variation in the World's Largest Ethnic Group —Han Chinese". Molecular Biology and Evolution. 35 (11): 2736–2750. doi:10.1093/molbev/msy170. PMC 6693441. PMID 30169787.
- He, Guang-Lin; Wang, Meng-Ge; Li, Ying-Xiang; et al. (2022). "Fine-scale north-to-south genetic admixture profile in Shaanxi Han Chinese revealed by genome-wide demographic history reconstruction". Journal of Systematics and Evolution. 60 (4): 955–972. doi:10.1111/jse.12715 – via Wiley Online Library.
- Zhou, Jingbin; Zhang, Xianpeng; Li, Xin; et al. (2022). "Genetic structure and demographic history of Northern Han people in Liaoning Province inferred from genome-wide array data". Frontiers in Ecology and Evolution. 10. doi:10.3389/fevo.2022.1014024.
- Sun, Na; Ma, Peng-Cheng; Yan, Shi; et al. (2019). "Phylogeography of Y-chromosome haplogroup Q1a1a-M120, a paternal lineage connecting populations in Siberia and East Asia". Annals of Human Biology. 46 (3): 261–266. doi:10.1080/03014460.2019.1632930. PMID 31208219 – via Taylor & Francis Online.
- Hurles, M; Sykes, B; Jobling, M; Forster, P (2005). "The Dual Origin of the Malagasy in Island Southeast Asia and East Africa: Evidence from Maternal and Paternal Lineages". The American Journal of Human Genetics. 76 (5): 894–901. doi:10.1086/430051. PMC 1199379. PMID 15793703.
- ^ He, Guanglin; Wang, Mengge; Miao, Lei; Chen, Jing; Zhao, Jie; Sun, Qiuxia; Duan, Shuhan; Wang, Zhiyong; Xu, Xiaofei; Sun, Yuntao; Liu, Yan; Liu, Jing; Wang, Zheng; Wei, Lanhai; Liu, Chao; Ye, Jian; Wang, Le (28 March 2023). "Multiple founding paternal lineages inferred from the newly-developed 639-plex Y-SNP panel suggested the complex admixture and migration history of Chinese people". Human Genomics. 17 (1): 29. doi:10.1186/s40246-023-00476-6. PMC 10045532. PMID 36973821.
- ^ Lu, Chuncheng; Zhang, Jie; Li, Yingchun; Xia, Yankai; Zhang, Feng; Wu, Bin; Wu, Wei; Ji, Guixiang; Gu, Aihua; Wang, Shoulin; Jin, Li; Wang, Xinru (2009). "The b2/b3 subdeletion shows higher risk of spermatogenic failure and higher frequency of complete AZFc deletion than the gr/gr subdeletion in a Chinese population". Human Molecular Genetics. 18 (6): 1122–30. doi:10.1093/hmg/ddn427. PMID 19088127.
- ^ Wen, B.; Li, H.; Lu, D.; Song, X.; Zhang, F.; He, Y.; Li, F.; Gao, Y.; et al. (September 2004). "Genetic evidence supports demic diffusion of Han culture" (PDF). Nature. 431 (7006): 302–05. Bibcode:2004Natur.431..302W. doi:10.1038/nature02878. PMID 15372031. S2CID 4301581. Archived from the original (PDF) on 24 March 2009.
- Xue, Fuzhong; Wang, Yi; Xu, Shuhua; Zhang, Feng; Wen, Bo; Wu, Xuesen; Lu, Ming; Deka, Ranjan; Qian, Ji; et al. (2008). "A spatial analysis of genetic structure of human populations in China reveals distinct difference between maternal and paternal lineages". European Journal of Human Genetics. 16 (6): 705–17. doi:10.1038/sj.ejhg.5201998. PMID 18212820.
- Wen, Bo; Li, Hui; Lu, Daru; Song, Xiufeng; Zhang, Feng; He, Yungang; Li, Feng; Gao, Yang; Mao, Xianyun; et al. (2004). "Genetic evidence supports demic diffusion of Han culture". Nature. 431 (7006): 302–05. Bibcode:2004Natur.431..302W. doi:10.1038/nature02878. PMID 15372031. S2CID 4301581.
- Chen, Jieming; Zheng, Houfeng; Bei, Jin-Xin; Sun, Liangdan; Jia, Wei-hua; Li, Tao; Zhang, Furen; Seielstad, Mark; Zeng, Yi-Xin; et al. (2009). "Genetic Structure of the Han Chinese Population Revealed by Genome-wide SNP Variation". The American Journal of Human Genetics. 85 (6): 775–85. doi:10.1016/j.ajhg.2009.10.016. PMC 2790583. PMID 19944401.
- McFadzean A.J.S., Todd D. (1971). "Cooley's anaemia among the tanka of South China". Transactions of the Royal Society of Tropical Medicine and Hygiene. 65 (1): 59–62. doi:10.1016/0035-9203(71)90185-4. PMID 5092429.
- Gan, Rui-Jing; Pan, Shang-Ling; Mustavich, Laura F.; Qin, Zhen-Dong; Cai, Xiao-Yun; Qian, Ji; Liu, Cheng-Wu; Peng, Jun-Hua; Li, Shi-Lin; Xu, Jie-Shun; Jin, Li; Li, Hui (2008). "Pinghua population as an exception of Han Chinese's coherent genetic structure". Journal of Human Genetics. 53 (4): 303–13. doi:10.1007/s10038-008-0250-x. PMID 18270655.
- Yang, Xiaomin; Wang, Xiao-Xun; He, Guanglin; et al. (2020). "Genomic insight into the population history of Central Han Chinese". Annals of Human Biology – via ResearchGate.
- Wang Y, Lu D, Chung YJ, Xu S (6 April 2018). "Genetic structure, divergence and admixture of Han Chinese, Japanese and Korean populations". Hereditas. 155 (1): 19. doi:10.1186/s41065-018-0057-5. PMC 5889524. PMID 29636655. *Xu S (10 April 2018). "Common ancestor of Han Chinese, Japanese and Koreans dated to 3000 – 3600 years ago". Biomed Central.
- Cao Y, Li L, Xu M, Feng Z, Sun X, Lu J, et al. (September 2020). "The ChinaMAP analytics of deep whole genome sequences in 10,588 individuals". Cell Research. 30 (9): 717–731. doi:10.1038/s41422-020-0322-9. PMC 7609296. PMID 32355288.
- ^ Zhao YB, Zhang Y, Zhang QC, Li HJ, Cui YQ, Xu Z, Jin L, Zhou H, Zhu H (2015). "Ancient DNA reveals that the genetic structure of the northern Han Chinese was shaped prior to 3,000 years ago". PLOS ONE. 10 (5): e0125676. Bibcode:2015PLoSO..1025676Z. doi:10.1371/journal.pone.0125676. PMC 4418768. PMID 25938511.
- Li J, Zeng W, Zhang Y, Ko AM, Li C, Zhu H, Fu Q, Zhou H (December 2017). "Ancient DNA reveals genetic connections between early Di-Qiang and Han Chinese". BMC Evolutionary Biology. 17 (1): 239. Bibcode:2017BMCEE..17..239L. doi:10.1186/s12862-017-1082-0. PMC 5716020. PMID 29202706.
- Yang MA, Fan X, Sun B, Chen C, Lang J, Ko YC, et al. (July 2020). "Ancient DNA indicates human population shifts and admixture in northern and southern China". Science. 369 (6501): 282–288. Bibcode:2020Sci...369..282Y. doi:10.1126/science.aba0909. PMID 32409524. S2CID 218649510.
- ^ Wen B, Li H, Lu D, Song X, Zhang F, He Y, et al. (September 2004). "Genetic evidence supports demic diffusion of Han culture". Nature. 431 (7006): 302–305. Bibcode:2004Natur.431..302W. doi:10.1038/nature02878. PMID 15372031. S2CID 4301581.
- Du R, Xiao C, Cavalli-Sforza LL (December 1997). "Genetic distances between Chinese populations calculated on gene frequencies of 38 loci". Science in China Series C: Life Sciences. 40 (6): 613–21. doi:10.1007/BF02882691. PMID 18726285. S2CID 1924085.
- Qin, Pengfei; Zhou, Ying; Lou, Haiyi; Lu, Dongsheng; Yang, Xiong; Wang, Yuchen; Jin, Li; Chung, Yeun-Jun; Xu, Shuhua (2 April 2015). "Quantitating and Dating Recent Gene Flow between European and East Asian Populations". Scientific Reports. 5 (1): 9500. Bibcode:2015NatSR...5.9500Q. doi:10.1038/srep09500. ISSN 2045-2322. PMC 4382708. PMID 25833680.
- Chen J, Zheng H, Bei JX, Sun L, Jia WH, Li T, et al. (December 2009). "Genetic structure of the Han Chinese population revealed by genome-wide SNP variation". American Journal of Human Genetics. 85 (6): 775–785. doi:10.1016/j.ajhg.2009.10.016. PMC 2790583. PMID 19944401.
- Gan RJ, Pan SL, Mustavich LF, Qin ZD, Cai XY, Qian J, Liu CW, Peng JH, Li SL, Xu JS, Jin L, Li H (2008). "Pinghua population as an exception of Han Chinese's coherent genetic structure". Journal of Human Genetics. 53 (4): 303–13. doi:10.1007/s10038-008-0250-x. PMID 18270655.
- ^ He, Guanglin; Wang, Mengge; Miao, Lei; Chen, Jing; Zhao, Jie; Sun, Qiuxia; Duan, Shuhan; Wang, Zhiyong; Xu, Xiaofei; Sun, Yuntao; Liu, Yan; Liu, Jing; Wang, Zheng; Wei, Lanhai; Liu, Chao (28 March 2023). "Multiple founding paternal lineages inferred from the newly-developed 639-plex Y-SNP panel suggested the complex admixture and migration history of Chinese people". Human Genomics. 17 (1): 29. doi:10.1186/s40246-023-00476-6. ISSN 1479-7364. PMC 10045532. PMID 36973821.
- Yao YG, Kong QP, Bandelt HJ, Kivisild T, Zhang YP (March 2002). "Phylogeographic differentiation of mitochondrial DNA in Han Chinese". American Journal of Human Genetics. 70 (3): 635–51. doi:10.1086/338999. PMC 384943. PMID 11836649.
- Kivisild T, Tolk HV, Parik J, Wang Y, Papiha SS, Bandelt HJ, Villems R (October 2002). "The emerging limbs and twigs of the East Asian mtDNA tree". Molecular Biology and Evolution. 19 (10): 1737–51. doi:10.1093/oxfordjournals.molbev.a003996. PMID 12270900.
- Yao YG, Kong QP, Man XY, Bandelt HJ, Zhang YP (February 2003). "Reconstructing the evolutionary history of China: a caveat about inferences drawn from ancient DNA". Molecular Biology and Evolution. 20 (2): 214–9. doi:10.1093/molbev/msg026. PMID 12598688.
- Kong QP, Sun C, Wang HW, Zhao M, Wang WZ, Zhong L, Hao XD, Pan H, Wang SY, Cheng YT, Zhu CL, Wu SF, Liu LN, Jin JQ, Yao YG, Zhang YP (January 2011). "Large-scale mtDNA screening reveals a surprising matrilineal complexity in east Asia and its implications to the peopling of the region". Molecular Biology and Evolution. 28 (1): 513–22. doi:10.1093/molbev/msq219. PMID 20713468.
- Liu, Chi-Chun; Witonsky, David; Gosling, Anna; Lee, Ju Hyeon; Ringbauer, Harald; Hagan, Richard; Patel, Nisha; Stahl, Raphaela; Novembre, John; Aldenderfer, Mark; Warinner, Christina; Di Rienzo, Anna; Jeong, Choongwon (8 March 2022). "Ancient genomes from the Himalayas illuminate the genetic history of Tibetans and their Tibeto-Burman speaking neighbors". Nature Communications. 13 (1): 1203. Bibcode:2022NatCo..13.1203L. doi:10.1038/s41467-022-28827-2. ISSN 2041-1723. PMC 8904508. PMID 35260549. S2CID 247317520.
- Wang, Hongru; Yang, Melinda A.; Wangdue, Shargan; Lu, Hongliang; Chen, Honghai; Li, Linhui; Dong, Guanghui; Tsring, Tinley; Yuan, Haibing; He, Wei; Ding, Manyu; Wu, Xiaohong; Li, Shuai; Tashi, Norbu; Yang, Tsho (15 March 2023). "Human genetic history on the Tibetan Plateau in the past 5100 years". Science Advances. 9 (11): eadd5582. Bibcode:2023SciA....9D5582W. doi:10.1126/sciadv.add5582. ISSN 2375-2548. PMC 10022901. PMID 36930720.
- Bhandari, Sushil; Zhang, Xiaoming (5 November 2015). "Genetic evidence of a recent Tibetan ancestry to Sherpas in the Himalayan region". Scientific Reports. 5: 16249. Bibcode:2015NatSR...516249B. doi:10.1038/srep16249. ISSN 2045-2322. PMC 4633682. PMID 26538459. "Comparing Sherpas, Tibetans, and Han Chinese showed that the D-M174 is the predominant haplogroup in Sherpas (43.38%) and prevalent in Tibetans (52.84%)5, but rare among both Han Chinese (1.4–6.51%)6,7 and other Asian populations (0.02–0.07%)8, aside from Japanese (34.7%) who possesses a distinct D-M174 lineage highly diverged from those in Tibetans and other Asian populations9,10."
- Zhang, Ganyu; Cui, Can; Wangdue, Shargan (16 March 2023). "Maternal genetic history of ancient Tibetans over the past 4000 years". Journal of Genetics and Genomics. 50 (10): 765–775. doi:10.1016/j.jgg.2023.03.007. PMID 36933795. S2CID 257588399.
- Blench R, Spriggs M (2 September 2003). Archaeology and Language II: Archaeological Data and Linguistic Hypotheses. Routledge. ISBN 978-1-134-82869-2.
- Yunusbayev, Bayazit; Metspalu, Mait; Metspalu, Ene; Valeev, Albert; Litvinov, Sergei; Valiev, Ruslan; Akhmetova, Vita; Balanovska, Elena; Balanovsky, Oleg; Turdikulova, Shahlo; Dalimova, Dilbar; Nymadawa, Pagbajabyn; Bahmanimehr, Ardeshir; Sahakyan, Hovhannes; Tambets, Kristiina (21 April 2015). "The Genetic Legacy of the Expansion of Turkic-Speaking Nomads across Eurasia". PLOS Genetics. 11 (4): e1005068. doi:10.1371/journal.pgen.1005068. ISSN 1553-7404. PMC 4405460. PMID 25898006.
- Nelson S, Zhushchikhovskaya I, Li T, Hudson M, Robbeets M (2020). "Tracing population movements in ancient East Asia through the linguistics and archaeology of textile production". Evolutionary Human Sciences. 2: e5. doi:10.1017/ehs.2020.4. ISSN 2513-843X. PMC 10427276. PMID 37588355. S2CID 213436897.
- Li T, Ning C, Zhushchikhovskaya IS, Hudson MJ, Robbeets M (1 June 2020). "Millet agriculture dispersed from Northeast China to the Russian Far East: Integrating archaeology, genetics, and linguistics". Archaeological Research in Asia. 22: 100177. doi:10.1016/j.ara.2020.100177. hdl:21.11116/0000-0005-D82B-8. ISSN 2352-2267. S2CID 213952845.
- Uchiyama J, Gillam JC, Savelyev A, Ning C (2020). "Populations dynamics in Northern Eurasian forests: a long-term perspective from Northeast Asia". Evolutionary Human Sciences. 2: e16. doi:10.1017/ehs.2020.11. ISSN 2513-843X. PMC 10427466. PMID 37588381. S2CID 219470000.
- Golden, Peter B. (October 2018). "The Ethnogonic Tales of the Türks". The Medieval History Journal. 21 (2): 291–327. doi:10.1177/0971945818775373. ISSN 0971-9458. S2CID 166026934.
- Dai, Shan-Shan; Sulaiman, Xierzhatijiang; Isakova, Jainagul; Xu, Wei-Fang; Abdulloevich, Najmudinov Tojiddin; Afanasevna, Manilova Elena; Ibrohimovich, Khudoidodov Behruz; Chen, Xi; Yang, Wei-Kang; Wang, Ming-Shan; Shen, Quan-Kuan; Yang, Xing-Yan; Yao, Yong-Gang; Aldashev, Almaz A; Saidov, Abdusattor (25 August 2022). "The Genetic Echo of the Tarim Mummies in Modern Central Asians". Molecular Biology and Evolution. 39 (9). doi:10.1093/molbev/msac179. ISSN 0737-4038. PMC 9469894. PMID 36006373.
- Guarino-Vignon, Perle; Marchi, Nina; Bendezu-Sarmiento, Julio; Heyer, Evelyne; Bon, Céline (14 January 2022). "Genetic continuity of Indo-Iranian speakers since the Iron Age in southern Central Asia". Scientific Reports. 12 (1): 733. Bibcode:2022NatSR..12..733G. doi:10.1038/s41598-021-04144-4. ISSN 2045-2322. PMC 8760286. PMID 35031610.
- Zerjal T, Wells RS, Yuldasheva N, Ruzibakiev R, Tyler-Smith C (September 2002). "A genetic landscape reshaped by recent events: Y-chromosomal insights into central Asia". American Journal of Human Genetics. 71 (3): 466–82. doi:10.1086/342096. PMC 419996. PMID 12145751.
- Kumar D (11 May 2012). Genomics and Health in the Developing World. Oxford, England: Oxford University Press. pp. 1265–1267. ISBN 978-0-19-970547-4.
- Damgaard PB, Marchi N, Rasmussen S, Peyrot M, Renaud G, Korneliussen T, et al. (May 2018). "137 ancient human genomes from across the Eurasian steppes". Nature. 557 (7705): 369–374. Bibcode:2018Natur.557..369D. doi:10.1038/s41586-018-0094-2. hdl:1887/3202709. PMID 29743675. S2CID 13670282.
- Yunusbayev B, Metspalu M, Metspalu E, Valeev A, Litvinov S, Valiev R, et al. (April 2015). "The genetic legacy of the expansion of Turkic-speaking nomads across Eurasia". PLOS Genetics. 11 (4): e1005068. doi:10.1371/journal.pgen.1005068. PMC 4405460. PMID 25898006.
- Meltzer DJ (2021). First Peoples in a New World: Populating Ice Age America. Cambridge University Press. p. 170. ISBN 978-1-108-49822-7.
- Meltzer 2021, p. 170: "The ancestors of Native Americans derive from descendants of Ancient North Siberians and Ancient North Eurasians on the one hand, (exemplified by the genomes of the Yana and Mal'ta individuals, respectively), and a population of Ancient East Asians (signified by the Tianyuan individual). From those groups emerged the deepest ancestors of Native Americans, who became separated far in far northeastern Asia from their Siberian and Asian ancestors around the time of the LGM. Part of the population stayed in Siberia (the Ancient Paleo-Siberians, seen in the Kolyma genome), while others moved east across Beringia."
- Lazaridis et al. 2018.
- Moreno-Mayar et al. 2018.
- Yang MA, Gao X, Theunert C, Tong H, Aximu-Petri A, Nickel B, et al. (October 2017). "40,000-Year-Old Individual from Asia Provides Insight into Early Population Structure in Eurasia". Current Biology. 27 (20): 3202–3208.e9. Bibcode:2017CBio...27E3202Y. doi:10.1016/j.cub.2017.09.030. PMC 6592271. PMID 29033327.
- Kidd KK, Evsanaa B, Togtokh A, Brissenden JE, Roscoe JM, Dogan M, et al. (May 2022). "North Asian population relationships in a global context". Scientific Reports. 12 (1): 7214. Bibcode:2022NatSR..12.7214K. doi:10.1038/s41598-022-10706-x. PMC 9068624. PMID 35508562.
- ^ Moreno-Mayar JV, Potter BA, Vinner L, Steinrücken M, Rasmussen S, Terhorst J, et al. (January 2018). "Terminal Pleistocene Alaskan genome reveals first founding population of Native Americans" (PDF). Nature. 553 (7687): 203–207. Bibcode:2018Natur.553..203M. doi:10.1038/nature25173. PMID 29323294. S2CID 4454580.
- Davis LG, Madsen DB, Becerra-Valdivia L, Higham T, Sisson DA, Skinner SM, et al. (August 2019). "Late Upper Paleolithic occupation at Cooper's Ferry, Idaho, USA, ~16,000 years ago". Science. 365 (6456): 891–897. Bibcode:2019Sci...365..891D. doi:10.1126/science.aax9830. PMID 31467216.
- Posth C, Nakatsuka N, Lazaridis I, Skoglund P, Mallick S, Lamnidis TC, et al. (November 2018). "Reconstructing the Deep Population History of Central and South America". Cell. 175 (5): 1185–1197.e22. doi:10.1016/j.cell.2018.10.027. PMC 6327247. PMID 30415837.
- Willerslev E, Meltzer DJ (June 2021). "Peopling of the Americas as inferred from ancient genomics". Nature. 594 (7863): 356–364. Bibcode:2021Natur.594..356W. doi:10.1038/s41586-021-03499-y. PMID 34135521. S2CID 235460793.
- Sarkar AA (18 June 2021). "Ancient Human Genomes Reveal Peopling of the Americas". GEN – Genetic Engineering and Biotechnology News. Retrieved 15 September 2021.
The team discovered that the Spirit Cave remains came from a Native American while dismissing a longstanding theory that a group called Paleoamericans existed in North America before Native Americans.
- Yelmen B, Mondal M, Marnetto D, Pathak AK, Montinaro F, Gallego Romero I, et al. (August 2019). "Ancestry-Specific Analyses Reveal Differential Demographic Histories and Opposite Selective Pressures in Modern South Asian Populations". Molecular Biology and Evolution. 36 (8): 1628–1642. doi:10.1093/molbev/msz037. PMC 6657728. PMID 30952160.
- ^ Chaubey G (January 2015). "East Asian ancestry in India" (PDF). Indian Journal of Physical Anthropology and Human Genetics. 34 (2): 193–199.
Here the analysis of genome wide data on Indian and East/Southeast Asian demonstrated their restricted distinctive ancestry in India mainly running along the foothills of Himalaya and northeastern part.
- ^ Chaubey G, Metspalu M, Choi Y, Mägi R, Romero IG, Soares P, et al. (February 2011). "Population genetic structure in Indian Austroasiatic speakers: the role of landscape barriers and sex-specific admixture". Molecular Biology and Evolution. 28 (2): 1013–1024. doi:10.1093/molbev/msq288. PMC 3355372. PMID 20978040.
- ^ Zhang X, Liao S, Qi X, Liu J, Kampuansai J, Zhang H, Yang Z, Serey B, Sovannary T, Bunnath L, Seang Aun H, Samnom H, Kangwanpong D, Shi H, Su B (October 2015). "Y-chromosome diversity suggests southern origin and Paleolithic backwave migration of Austro-Asiatic speakers from eastern Asia to the Indian subcontinent". Scientific Reports. 5: 15486. Bibcode:2015NatSR...515486Z. doi:10.1038/srep15486. PMC 4611482. PMID 26482917.
- ^ Ness I (2014). The Global Prehistory of Human Migration. Chichester, West Sussex: John Wiley & Sons. p. 265. ISBN 978-1-118-97058-4.
- ^ Chaubey G, Metspalu M, Choi Y, Mägi R, Romero IG, Soares P, et al. (February 2011). "Population genetic structure in Indian Austroasiatic speakers: the role of landscape barriers and sex-specific admixture". Molecular Biology and Evolution. 28 (2): 1013–1024. doi:10.1093/molbev/msq288. PMC 3355372. PMID 20978040.
- ^ Van Driem G (2007). "Austroasiatic phylogeny and the Austroasiatic homeland in light of recent population genetic studies" (PDF). Mon-Khmer Studies. 37: 1–14.
- ^ Riccio ME, Nunes JM, Rahal M, Kervaire B, Tiercy JM, Sanchez-Mazas A (June 2011). "The Austroasiatic Munda population from India and Its enigmatic origin: a HLA diversity study". Human Biology. 83 (3): 405–35. doi:10.3378/027.083.0306. PMID 21740156. S2CID 39428816.
- ^ Arunkumar G, Wei LH, Kavitha VJ, Syama A, Arun VS, Sathua S, et al. (November 2015). "A late Neolithic expansion of Y chromosomal haplogroup O2a1-M95 from east to west: Late Neolithic expansion of O2a1-M95". Journal of Systematics and Evolution. 53 (6): 546–560. doi:10.1111/jse.12147. S2CID 83103649.
- "ASI-AAA"
- Valar M (21 April 2015). "DNA Reveals Unknown Ancient Migration Into India". National Geographic. Archived from the original on 11 June 2015.
- Gutman A, Avanzati B. "Austroasiatic Languages". The Language Gulper.
- Yelmen B, Mondal M, Marnetto D, Pathak AK, Montinaro F, Gallego Romero I, et al. (August 2019). "Ancestry-Specific Analyses Reveal Differential Demographic Histories and Opposite Selective Pressures in Modern South Asian Populations". Molecular Biology and Evolution. 36 (8): 1628–1642. doi:10.1093/molbev/msz037. PMC 6657728. PMID 30952160.
- Lévi S, Przyluski J, Bloch J (1993). Pre-Aryan and Pre-Dravidian in India. Asian Educational Services. ISBN 978-81-206-0772-9.
It has been further proved that not only linguistic but also certain cultural and political facts of ancient India, can be explained by Austroasiatic elements.
- "How rice farming may have spread across the ancient world". science.org. Retrieved 26 October 2021.
- Changmai P, Pinhasi R, Pietrusewsky M, Stark MT, Ikehara-Quebral RM, Reich D, Flegontov P (December 2022). "Ancient DNA from Protohistoric Period Cambodia indicates that South Asians admixed with local populations as early as 1st-3rd centuries CE". Scientific Reports. 12 (1): 22507. Bibcode:2022NatSR..1222507C. doi:10.1038/s41598-022-26799-3. PMC 9800559. PMID 36581666.
- Liu D, Duong NT, Ton ND, Van Phong N, Pakendorf B, Van Hai N, Stoneking M (September 2020). "Extensive Ethnolinguistic Diversity in Vietnam Reflects Multiple Sources of Genetic Diversity". Molecular Biology and Evolution. 37 (9): 2503–2519. doi:10.1093/molbev/msaa099. PMC 7475039. PMID 32344428.
- Carlhoff S, Duli A, Nägele K, Nur M, Skov L, Sumantri I, et al. (August 2021). "Genome of a middle Holocene hunter-gatherer from Wallacea". Nature. 596 (7873): 543–547. Bibcode:2021Natur.596..543C. doi:10.1038/s41586-021-03823-6. hdl:10072/407535. PMC 8387238. PMID 34433944.
The qpGraph analysis confirmed this branching pattern, with the Leang Panninge individual branching off from the Near Oceanian clade after the Denisovan gene flow, although with the most supported topology indicating around 50% of a basal East Asian component contributing to the Leang Panninge genome (Fig. 3c, Supplementary Figs. 7–11).
- Changmai P, Jaisamut K, Kampuansai J, Kutanan W, Altınışık NE, Flegontova O, et al. (February 2022). "Indian genetic heritage in Southeast Asian populations". PLOS Genetics. 18 (2): e1010036. doi:10.1371/journal.pgen.1010036. PMC 8853555. PMID 35176016.
Works cited
- Lazaridis I, Belfer-Cohen A, Mallick S, Patterson N, Cheronet O, Rohland N, et al. (21 September 2018). "Paleolithic DNA from the Caucasus reveals core of West Eurasian ancestry". bioRxiv 10.1101/423079.
- Lee J, Miller BK, Bayarsaikhan J, Johannesson E, Ventresca Miller A, Warinner C, Jeong C (April 2023). "Genetic population structure of the Xiongnu Empire at imperial and local scales". Science Advances. 9 (15): eadf3904. Bibcode:2023SciA....9F3904L. doi:10.1126/sciadv.adf3904. PMC 10104459. PMID 37058560.
- Li J, Zhang Y, Zhao Y, Chen Y, Ochir A, Zhu H, Zhou H (August 2018). "The genome of an ancient Rouran individual reveals an important paternal lineage in the Donghu population". American Journal of Physical Anthropology. 166 (4). American Association of Physical Anthropologists: 895–905. doi:10.1002/ajpa.23491. PMID 29681138.
Further reading
- He G, Wang Z, Guo J, Wang M, Zou X, Tang R, et al. (August 2020). "Inferring the population history of Tai-Kadai-speaking people and southernmost Han Chinese on Hainan Island by genome-wide array genotyping". European Journal of Human Genetics. 28 (8): 1111–1123. doi:10.1038/s41431-020-0599-7. PMC 7381617. PMID 32123326. S2CID 211729663.
| Human genetics | |
|---|---|
| Sub-topics | |
| Genetic history by region | |
| Population genetics by group |
|