| Revision as of 12:06, 22 July 2009 editSmall Victory (talk | contribs)412 edits →Editors showing their true colors← Previous edit | Revision as of 12:18, 22 July 2009 edit undoSmall Victory (talk | contribs)412 edits →go it alone editingNext edit → | ||
| Line 766: | Line 766: | ||
| Small Victory, concernint , just to remind you again, the deleted article was entirely your baby. You rejected contributions from myself and compromises from Andrew. The community rejected your approach with ]. That was essentially a referendum on your approach. So this time, we have to use another approach. ] was blocked yesterday for recreating the article ], what you are doing comes awfully close to what SOPHIAN is trying to do. I suggest you accept that you must work with others to achieve a consensus. ] (]) 14:11, 21 July 2009 (UTC) | Small Victory, concernint , just to remind you again, the deleted article was entirely your baby. You rejected contributions from myself and compromises from Andrew. The community rejected your approach with ]. That was essentially a referendum on your approach. So this time, we have to use another approach. ] was blocked yesterday for recreating the article ], what you are doing comes awfully close to what SOPHIAN is trying to do. I suggest you accept that you must work with others to achieve a consensus. ] (]) 14:11, 21 July 2009 (UTC) | ||
| :You're quite delusional. That article was deleted because it was a ]. And your POV-pushing, original research, 3RR violations and sock puppets had more to do with it than anything I ever did. In fact, the article was problem-free until you (and Andrew Lancaster) came along and started tampering with it. Let's remember that you're the one who's been for repeated rule violations. My record is . So if anything, the deletion was a referendum on '''''your''''' approach. Take the hint. ---- ] (]) 12:18, 22 July 2009 (UTC) | |||
| ==Luigi Luca Cavalli-Sforza== | ==Luigi Luca Cavalli-Sforza== | ||
Revision as of 12:18, 22 July 2009
| This article has not yet been rated on Misplaced Pages's content assessment scale. It is of interest to the following WikiProjects: | ||||||||||||||||||
Template:WikiProject Genetics
Template:Wikiproject MCBPlease add the quality rating to the {{WikiProject banner shell}} template instead of this project banner. See WP:PIQA for details.
| ||||||||||||||||||
Norman/Germanic - Viking heritage - Language and Culture in Sicily.
My cousin through marriage David Neilson assumed my surname was biological ‘Raciti’. I am biological in fact a Caggegi. He told me that I needed to wait in line before I could consider my Norman/Germanic - Viking heritage.
He believes he is of Danish heritage (through his surname - Neilson). He has brown hair and brown eyes. I personally don't see it at all. My daughter Racheal has blonde hair and blue eyes - and is most likely to be of that area.
I have found the original form of my biological name to be of a 'North Sea Germanic language' of Norse Origin: 'Keggeg', specifically from the Ingaevones, Jastorf and Langobardic cultures that migrated into North Italy in the 6th and 7th centuries.
History tells us that there were significant Lombard (with their Gallo-Italic idiom) settlements in Randazzo, Sicily.
The Langobardi tribe could have been biologically very similar to The Cimbri (Danes) and The Frisii tribes.
The one thing I do know is that The Lombards through The Jastorf culture - were in locations in Sweden - were I find other 'Keggeg's.
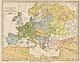
http://en.wikipedia.org/Image:Pre_Migration_Age_Germanic.png
New maps
Can someone integrate these maps into the article?: Phoenix of9 (talk) 00:15, 26 March 2009 (UTC)
- They're a bit hard to create, and I find them aesthetically displeasing Hxseek (talk) 07:07, 12 April 2009 (UTC)
Actually, that 2nd map is nice. I'll see if I can create anything similar. Looks a bit complicated Hxseek (talk) 23:08, 23 April 2009 (UTC)
Upgrade
I propose the following upgrade to improve the article (which I shall be happy to commence soon(ish). If any has further suggestions, then feel fre to add)
(1)To start off: discuss European's relationship to other continental populations, and to each other (as already done, but perhaps integrate more)
(2) Minimise the "haplogroups" section. Perhaps just a summary of the different modes of DNA evidence (autosomal, protien-level, Y, mtDNA), and which groups are found in Europe, the assumptions (and weaknesses) of genetic studies.
(3) Remove the western european substructure section (as well as the 2-lined eastern Euro section), as it is artifical to divide europe into east and west, at least geneticaly-speaking. This will be anyway covered in (1) above
(4) Keep the migrations section, but link it more with the genetic evidence. Add a paragraph on Neanderthals, and their (lack of) relationship with modern Europeans.
(5) We do not need to keep an entire section on "African influences" . Why does it deserve an entire paragraph for itself ? It can be worked into the rest of the article through the palaeolithic, neolithic , post-Roman migrations, etc, as supported by whatever evidence which might exist
Hxseek (talk) 10:08, 17 April 2009 (UTC)
- Agreed! Go for it! The Ogre (talk) 15:32, 17 April 2009 (UTC)
- Works for me too. — SMcCandlish ‹(-¿-)› 11:58, 13 May 2009 (UTC)
- I agree with most except, the African influences, shouldn't be spread out over several sections. Related information should be found in close proximity. Wapondaponda (talk) 15:28, 13 May 2009 (UTC)
- Works for me too. — SMcCandlish ‹(-¿-)› 11:58, 13 May 2009 (UTC)
Well, (to Wapondaponda) that's one way, sure. But then its not chronological. If we're talking of 'genetic history', shouldn;t we procees in some kind of temporal sequence ? Hxseek (talk) 00:05, 15 May 2009 (UTC)
The "percentage genetic distances" chart isn't helpful (yet)
The "percentage genetic distances" chart is not something that the average encyclopedia reader can understand. Even with a background in anthropology, I was only able to figure out what it was trying to convey after looking at it for quite a long time. The chart needs to be re-done in a much more intuitive fashion. — SMcCandlish ‹(-¿-)› 11:58, 13 May 2009 (UTC)
- Numerically, I think that current chart is probably the easiest way to express the information. The other possibility is to use a dendogram. Wapondaponda (talk) 15:20, 13 May 2009 (UTC)
Misleading inclusion of map & dishonest caption
I've removed a map one user has added (and not for the first time either) which, according to its caption, claims to indicate "the spread of haplogroup E-M78 from Northeast Africa". This map is sourced to a study by Cruciani et al. (2007) and claims to be based on that study's Figure 2A. However, Figure 2A of Cruciani et al. (2007) indicates the map is of "the observed haplogroup/paragroup frequencies." It says nothing about the "spread" of E-M78 much less from where that spread originated. In fact, Cruciani et al. (2007) indicate that E-V13, the most common sub-clade of E-M78 in Europe, originated in situ in the Balkans during the Bronze age. Its presence in Europe was not due to "recent medieval North African/Near Eastern admixture", (admixture which, incidentally, is usually restricted to E1b1b1b i.e. E-M81, not E-M78, the latter of which has an ancient presence in the region) so the map in question should not under any circumstances be placed in the section of the same name as the user has attempted. Moreover, a map of E-V13, the most common sub-clade of E1b1b found in Europe, has already been included in the article, and that map already accurately covers the distribution of E-V13 in Europe. Causteau (talk) 17:46, 14 May 2009 (UTC)
- Yes E-V13 originated in Europe, but the bearer of the mutation was someone who carried an E-M78 lineage that extended to Northeast Africa, and ultimately extended to east Africa. Along with his Y-chromosome, they certainly carried some sub-saharan dna, which is confirmed by Cavalli-Sforza's classical polymorphisms. With regard to M1, its origins are still disputed, but there is general agreement that M1a arose in East Africa, and this haplogroup is found in Europe too. Wapondaponda (talk) 18:05, 14 May 2009 (UTC)
- What a load of crock and an utter red herring:
- No kidding E-M78 originated in Northeast Africa (Egypt/Libya, for the unitiated); who said it didn't? E-M78 didn't just, however, "ultimately extend to East Africa". It spread from North Africa to the Horn, the Near East and Europe from its Egyptian homeland. East Africa doesn't ipso facto have any sort of "first dibs" on or "special relationship" with, as it were, the clade; it doesn't work that way.
- Cavalli-Sforza's work using classic autosomal markers asserts absolutely nothing of the sort you claim. You are quite literally fabricating information (and that has no bearing on the present article, to boot).
- No, the origin of M1 is not "disputed". Actually, it was settled quite a while back, as was the origin of its parent haplogroup M: Both haplogroups were accorded an Asian origin. But of course, you already know this since you've been trying for weeks now to manufacture a "debate" to revive the long-dead African origin hypothesis on the haplogroup M talk page. Now you bring up the origin of M1a, as if any modern study asserts that its parent M1 clade originated in East Africa. M1a isn't even the oldest sub-clade of M1, but of course you're busy attempting to deny that too on the haplogroup M talk page! LOL Causteau (talk) 18:25, 14 May 2009 (UTC)
E-M78 is a mutation of E-M215 for which there is no dispute of its East African origins. We could easily substitute E-M215 for E-M78 and it would be perfectly valid. Many Europeans today have a Y-Chromosome of an East African man. Wapondaponda (talk) 18:32, 14 May 2009 (UTC)
- That's absurd. E-M215 originated at least 30,000 year ago (and likely far before that, given Karafet et al. (2008)'s recent pushing back of haplogroup E's TMRCA to 52,500 years ago and E1b1's TMRCA to 47,500 ybp), a good 10,000 years+ before E-M78, which, by comparison, first arose only about 18,600 years ago. E-M215 also arose in a very different environment from both E-M78 & E-V13 with very different selective pressures acting upon the host population. I'd also add that R1a originated in Central Asia, yet it's the second most frequently observed haplogroup in Europe today. Using your logic, the many modern Europeans who carry the haplogroup today would therefore have "Asian" admixture, which is of course preposterous. Causteau (talk) 18:51, 14 May 2009 (UTC)
- Actually that is exactly what Cavalli-Sforza states He describes Europeans as being a mix of Africans and Asians. Wapondaponda (talk) 19:07, 14 May 2009 (UTC)
- Actually, that is not at all what Cavalli-Sforza states. He speaks of the genetic distance of Europeans between Sub-Saharan Africans and Asians as being a product of geographic distance, not admixture as you seem to be suggesting/hoping. Causteau (talk) 19:18, 14 May 2009 (UTC)
- Unless genes spread by air. The last I knew genes spread by sexual reproduction, and people tend to mate with people living nearby, ie geographic distance. He specifically states in the book, the article is an excerpt, that there has been significant gene flow between Europe and Africa in recent history and in prehistory. Hence Europeans occupy an intermediate position between sub-saharan Africa and Asia, by virtue of the classical polymorphisms. The y-chromosome is transmitted in whole, so out of 46 chromosomes, many europeans have one that is in essence of East African origin. Add to that the many other nuclear genes that must have come along with the Y chromosome, it makes sense. Cruciani et al estimate E-M215 to be 22000 years old. Wapondaponda (talk) 19:33, 14 May 2009 (UTC)
- Actually, that is not at all what Cavalli-Sforza states. He speaks of the genetic distance of Europeans between Sub-Saharan Africans and Asians as being a product of geographic distance, not admixture as you seem to be suggesting/hoping. Causteau (talk) 19:18, 14 May 2009 (UTC)
- Actually that is exactly what Cavalli-Sforza states He describes Europeans as being a mix of Africans and Asians. Wapondaponda (talk) 19:07, 14 May 2009 (UTC)
- Again, you're inventing stuff. Cavalli-Sforza make no such assertion which is why you are unable to quote him on the matter -- a matter which, incidentally, has absolutely nothing to do with your actual edits. What you are attempting to "prove" is that E1b1b-carrying Europeans are "in essence of East African origin", which is most amusing seeing as how the Y haplogroup tree does not stop with E-M215, as you had undoubtedly hoped. In actuality, it reaches back to E, DE, and beyond to a common paternal ancestor for all humans. So actually, channeling your logic again, E1b1b-carrying Europeans like all other humans are first and foremost Africans since that is where humanity as we know it ostensibly began. And if we observe where exactly orginated the most recent haplogroups said Europeans belong to, it's of course typically Europe. In the case of R1a, for example, it's Asia, but that again doesn't make said Europeans anymore "Asian" than your ridiculous assertion that E1b1b-carrying Europeans are "East African". Lastly, while Cruciani et al. (2007) did indeed say E-M215 originated 22,400 years ago, that estimate was from before Karafet et al. (2008)'s update of the Y tree. One can only imagine how far back E1b1b's TMRCA will get pushed back to now considering how far back its parent clade & haplogroup E's respective TMRCA were. Causteau (talk) 19:57, 14 May 2009 (UTC)
Misleading inclusion of map & dishonest caption section break 1
You guys always take the same sides! :) Anyway, let me go through a list of things I see:
- Frequencies, spread, distribution are the same. Causteau seems to think spread implies a direction of migration. I do not think so but anyway, Cruciani do imply such a direction in their text, so let's not go overboard.
- You are both wrong to say that Cruciani et al. say E-V13 originated in the Balkans. They say it dispersed from there. Battaglia et al. wonder, not too strongly, if it did indeed originate there, but I find that hard to see as mainstream given the way they argue this, or rather don't. Put it this way: Cruciani et al. make a strong case it originated in the Levant. (It did not get common maybe until much later in the Balkans.)
- I am not sure if Cavalli-Sforza's classic work used what we would today call autosomal markers. Not sure on this.
- I think Wapondaponda has mixed up E-M78 and E-M35. The current thinking is that E-M78 is much younger and has a more northerly origin than its parent E-M35 - somewhere around Egypt rather than somewhere around Ethiopia. E-M35 can be said to be equivalent to E-M215, not E-M78. OTOH I think Causteau's certainty about E-M215 being >30,000 is over-stated.
- I think it is dubious to say R1a is more common than I haplogroups and I've had discussions before with Causterau about the pointlessness of trying to compare how common different clades are which are not clearly siblings. It is not comparing apples with apples.
- Wapondaponda did not say that some Europeans were of East African origin, but rather their Y chromosome, which is true in some way. All of us are African, and all our genes. The question is how to say what he means. I think we need to handle this in a section beyond Cavalli-Sforza. The sections relating to the LATEST literature should acknowledge that there are still big questions about how many major emigrations there were from Africa, and when. I believe it is absolutely orthodox to say that, as Underhill said in a review not too long ago, E1b1b is a rare case of a relatively recent movement from Africa to Europe. Of course other genes went with it. I believe it is not going too far to say that there is a pretty big orthodoxy out there saying that this emigration was around about the time the Neolithic initiated.--Andrew Lancaster (talk) 20:32, 14 May 2009 (UTC)
- Thank you for your input, Andrew. But what you have responded to above is Wapondaponda's red herring "debate", not to his actual edits (which is the point of talk pages). The actual content of his edits are analysed in my first post in this section of the talk page (the one dated 17:46, 14 May 2009). Please respond to that. Causteau (talk) 20:42, 14 May 2009 (UTC)
- Cavalli-Sforza's work was primarily on amino acid sequences from blood proteins. So their studies are based on phenotype that is expressed from the underlying genotype. So yes it is a proxy for autosomal DNA, but much of his studies were before PCR. Nonetheless, current studies have basically confirmed much of Sforza's work. Wapondaponda (talk) 21:33, 14 May 2009 (UTC)
- Quotes from Cavalli Sforza
- Europe shows a shorter (genetic) distance from Africa than do all the other continents. The difference is statistically significant and is consistently found with all markers, ranging from “classical” ones based on gene products to DNA markers such as restriction polymorphisms (4) and microsatellites (5).
- Wapondaponda (talk) 21:53, 14 May 2009 (UTC)
- Cavalli-Sforza's work was primarily on amino acid sequences from blood proteins. So their studies are based on phenotype that is expressed from the underlying genotype. So yes it is a proxy for autosomal DNA, but much of his studies were before PCR. Nonetheless, current studies have basically confirmed much of Sforza's work. Wapondaponda (talk) 21:33, 14 May 2009 (UTC)
- Thank you for your input, Andrew. But what you have responded to above is Wapondaponda's red herring "debate", not to his actual edits (which is the point of talk pages). The actual content of his edits are analysed in my first post in this section of the talk page (the one dated 17:46, 14 May 2009). Please respond to that. Causteau (talk) 20:42, 14 May 2009 (UTC)
- Nuh... you don't say Europe is closer genetically to Africa than, say, East Asia is to Africa (a fact which isn't necessarily attributable to gene flow, by the way)? Thanks for pointing out the obvious, Wapondaponda. You conveniently forgot to mention, however, that Europe is closer genetically overall to all other continents than it is to Africa. It is just geographically close to Africa. That's the difference between two people sitting on one side of a long bench and another person sitting way on the other side of the same bench: One of the two in the nether regions is necessarily closer to the person all the way at the other end of the bench, but certainly not closer to that person than to the one right beside them. I don't suppose you've actually looked at the genetic distances involved have you? If you had, you'd know better than to concoct such a silly argument. It's like this: nowhere in the quote above does Cavalli-Sforza state that the people who introduced E-M78 to Europe had "some sub-saharan dna" as you originally claimed, 2) nowhere does it state that this supposed admixture "is confirmed by classical polymorphisms" as you have also claimed, and 3) nowhere does Cavalli-Sforza even come close to describing "Europeans as being a mix of Africans and Asians", which was the thrust of your digression. The forgoing is just more of that famous original research you can't seem to stop producing. Causteau (talk) 22:41, 14 May 2009 (UTC)
- The full details are in the book . Unfortunately it is not available online. Basically Europeans are the population with the most African admixture. A review from the nytimes states
Misleading inclusion of map & dishonest caption section break 2
- Cavalli-Sforza shows that the European population is the most genetically mixed-up on earth, being a mix of genes from Asia and Africa.GENES, PEOPLES,AND LANGUAGESWapondaponda (talk) 23:04, 14 May 2009 (UTC)
- Oh, I see. LOL So the full details are in the book now. See, I could've sworn you asserted (and rather confidently) not once but twice that the relevant information was in the article above. I could've sworn you linked me to the article in question and even quoted a passage directly from it which you believed supposedly "supported" your claims. But now I see that you are reduced to quoting lay journalists (a book reviewer, no less) rather than Cavalli-Sforza himself to support your absurd argument, an argument which wasn't, by the way, that "Europeans are the population with the most African admixture"; that's your new and necessarily less ambitious argument -- I wonder why that is? It's the book reviewer that says that Europeans are "a mix of genes from Asia and Africa". Cavalli-Sforza himself does not and you know it, which again is why you are unable to quote the relevant passages from the book despite claiming to have read it. That "it is not available online" is a cop-out, but an understandable one given the situation you are in. I'll tell you what though: Cite which exact section & page number of the book the famous information is located in, and I will only too happily look it up for the both of us and relay to you what we both already know (or perhaps you'll do the honors and quote it directly yourself?). Causteau (talk) 00:07, 15 May 2009 (UTC)
- page 52 but depends on the edition,
- In order to examine the problem of the constancy of evolutionary rates, we can look at the distances between Africa and the other continents: 24.7 with Oceania, 20.6 with Asia, 16.6 with Europe, and 22.6 with America. It is clear that the shortest distance is between Africa and Europe, followed by that between Africa and Asia. If the rate of evolution were truly constant, the four values would be identical (within the limits of statistical error due to small sample size)
- Oh, I see. LOL So the full details are in the book now. See, I could've sworn you asserted (and rather confidently) not once but twice that the relevant information was in the article above. I could've sworn you linked me to the article in question and even quoted a passage directly from it which you believed supposedly "supported" your claims. But now I see that you are reduced to quoting lay journalists (a book reviewer, no less) rather than Cavalli-Sforza himself to support your absurd argument, an argument which wasn't, by the way, that "Europeans are the population with the most African admixture"; that's your new and necessarily less ambitious argument -- I wonder why that is? It's the book reviewer that says that Europeans are "a mix of genes from Asia and Africa". Cavalli-Sforza himself does not and you know it, which again is why you are unable to quote the relevant passages from the book despite claiming to have read it. That "it is not available online" is a cop-out, but an understandable one given the situation you are in. I'll tell you what though: Cite which exact section & page number of the book the famous information is located in, and I will only too happily look it up for the both of us and relay to you what we both already know (or perhaps you'll do the honors and quote it directly yourself?). Causteau (talk) 00:07, 15 May 2009 (UTC)
- The distance from Europe is anomalously low. North Africa is populated with Caucasoid people like Europeans, but we have made sure to eliminate these populations and are restricting ourselves to Sub-Saharan Africa. The simplest explanation is that substantial exchange has taken place between nearby continents.
- Wapondaponda (talk) 00:23, 15 May 2009 (UTC)
- Thank you for producing that quote, Wapondaponda. Now that I see it, it's pretty clear why you seemed hesitant to produce it in the first place. The quote above directly contradicts your actual edits. You ostensibly brought up Cavalli-Sforza as a way of "proving" that E-M78 in Europe somehow constitutes "Sub-Saharan" admixture. Yet, there is Cavalli-Sforza grouping North Africans with Europeans as "Caucasoid people" and specifying that he and his team "made sure to eliminate these populations and are restricting ourselves to Sub-Saharan Africa." Remember that the section in question in the Genetic history of Europe article is on recent medieval North African/Near Eastern admixture in Europe -- not Sub-Saharan admixture. You have again shot yourself in the foot my friend, but the irony is that this never would have happened had you just let it go instead of pushing it as is your wont. Causteau (talk) 00:49, 15 May 2009 (UTC)
- Yes it does, if it originated in Africa fairly recently, and ended up in Europe, its admixture. Even when Cavalli-Sforza eliminated North Africans, and compared Europeans with Sub-Saharans, he found an anomalously short genetic distance. If there were no admixture between Europe and Africa, Europe should have had a genetic distance similar to that of Australia, at 24.7, rather Europe has a genetic distance from Africa of 16.6. True Cavalli-Sforza doesn't reference E-M78, but talks about autosomal admixture. Nonetheless, there is a correlation. As Andrew mentioned, most likely during the Neolithic, there was demic diffusion of farmers from the Near East into Europe. Due to proximity to Africa, some of the farmers who brought agriculture to Europe were had African admixture and carried E-M78 along with other sub-saharan nuclear markers into Europe. This is clearly illustrated in Cruciani's map or Semino et al. Of course the neolithic is just one event, in paleolithic times there would have been several migrations.
- Note that prior to the development of Agriculture, all humans were hunter gatherers, who lived an egalitarian lifestyle. The notion of European, Asian or African didn't apply to hunter gatherers. They went where the food was, and picked up mates wherever they were available. Specifically since in paleolithic times, population density was really low, hunter gatherers couldn't afford to bother whether someone was from originally from Asia, Africa or Europe as may be the case in the modern world. It shouldn't come as a surprise to anyone that Europeans have significant African admixture. Wapondaponda (talk) 02:18, 15 May 2009 (UTC)
- Thank you for producing that quote, Wapondaponda. Now that I see it, it's pretty clear why you seemed hesitant to produce it in the first place. The quote above directly contradicts your actual edits. You ostensibly brought up Cavalli-Sforza as a way of "proving" that E-M78 in Europe somehow constitutes "Sub-Saharan" admixture. Yet, there is Cavalli-Sforza grouping North Africans with Europeans as "Caucasoid people" and specifying that he and his team "made sure to eliminate these populations and are restricting ourselves to Sub-Saharan Africa." Remember that the section in question in the Genetic history of Europe article is on recent medieval North African/Near Eastern admixture in Europe -- not Sub-Saharan admixture. You have again shot yourself in the foot my friend, but the irony is that this never would have happened had you just let it go instead of pushing it as is your wont. Causteau (talk) 00:49, 15 May 2009 (UTC)
- Your original research is getting very annoying. Cavalli-Sforza assert no such thing, which is why you are still unable to produce a quote supporting your claims. The shorter genetic distance of Europe from Africa compared to the genetic distance of other continents from Africa is not necessarily caused by admixture/gene flow. This is what you personally would like it to be, but haven't produced any evidence supporting that when clearly asked to. In case you are wondering, evidence means quotes directly from Cavalli-Sforza, the reliable source in question per WP:PROVEIT. You have been told by two separate editors now -- myself and Hxseek -- that the recent admixture in Europe involved E-M81, not the E-M78 indicated in that map based on Cruciani et al. (2007) that you've been attempting to add to the article. According to Battaglia et al. (2008), E-M78 in the form of its dominant E-V13 sub-clade represents ancient in situ differentiation. Cruciani suggests that perhaps it may have arisen in Anatolia, which is hardly Sub-Saharan Africa, making your claims of "Sub-Saharan admixture" all the more preposterous. And even prior to that, when E-M78 first arose in North Africa, according to the Cavalli-Sforza quote you yourself produced, that North Africa is populated by "Caucasoid people like Europeans", not Sub-Saharan Africans. You write that "due to proximity to Africa, some of the farmers who brought agriculture to Europe were had African admixture and carried E-M78 along with other sub-saharan nuclear markers into Europe." What's amusing about this quote is that E-M78 is not Sub-Saharan. It is Northeast African (i.e. Egypt/Libya), as Cruciani himself has made clear in his 2007 study. You also claim that that quote "is clearly illustrated in Cruciani's map or Semino et al." Ahhh, no it isn't. Again:
Causteau (talk) 11:14, 15 May 2009 (UTC)I've removed a map one user has added (and not for the first time either) which, according to its caption, claims to indicate "the spread of haplogroup E-M78 from Northeast Africa". This map is sourced to a study by Cruciani et al. (2007) and claims to be based on that study's Figure 2A. However, Figure 2A of Cruciani et al. (2007) indicates the map is of "the observed haplogroup/paragroup frequencies." It says nothing about the "spread" of E-M78 much less from where that spread originated. In fact, Cruciani et al. (2007) indicate that E-V13, the most common sub-clade of E-M78 in Europe, originated in situ in the Balkans during the Bronze age. Its presence in Europe was not due to "recent medieval North African/Near Eastern admixture", (admixture which, incidentally, is usually restricted to E1b1b1b i.e. E-M81, not E-M78, the latter of which has an ancient presence in the region) so the map in question should not under any circumstances be placed in the section of the same name as the user has attempted. Moreover, a map of E-V13, the most common sub-clade of E1b1b found in Europe, has already been included in the article, and that map already accurately covers the distribution of E-V13 in Europe.
Misleading inclusion of map & dishonest caption section break 3
I've got a hard copy of Cavalli-Sforza's book. I'll check what he exactly says about the mixing of Europe.
As for E M78, from what I know, it originated somewhere in northeastern Africa. Battaglia proposes that it spread northward by foragers during the increased precipation period associated with the Holocene. No-one is sure exactly where V13 itself arose. Obviusly, as these foragers dispersed, new mutations arose. Cruciani alludes to a possible Anatolian origin, then moving into the Balkans anytime after 17 kya. he connects its actual expansion with the Balkan Bronze Age. Instead, Battaglia and King link it to late Mesolithic/ early Neloithic expansions from within the Balkans also, tentatively placing its origin to somewhere in the southern balkans. With such uncertainty, and wide confidence intervals, we can hardly embrace any hypothesis. (this is the problem with genetic studies).
None of this apparent African madmixture is reflected in Cavalli-Sforza's discussion of principal components. I.e. none of his 5 PCs show a cline emanating from northern Africa. Makes sense, given that Y DNA data do not show any significant direct migration from Africa, except a few, different lineages such as M81, which are limited to Iberia and southern Italy. Instead Y DNA data point to central Eurasian and western Eurasian origins of current European Y DNA pool, in the form of the R1 and IJ supercomplexes, respectively {and I use "Eurasian" because, geographically, Europe and Asia are one continent, and their separation is political/ cultural. Therefore we do not need to speak of "Asian" origins, and the emotive responses it might elicit in some readers). Hxseek (talk) 00:27, 15 May 2009 (UTC)
- Actually the first PC is cline emanating from the Near East indicating the spread of farming. It is this cline that would be representative of African admixture as populations from the Near East had some African influences. Wapondaponda (talk) 00:50, 16 May 2009 (UTC)
- Thanks for your input, Hxseek. I think that's a fair summary of the situation. Causteau (talk) 00:49, 15 May 2009 (UTC)
- I also think Hxseek's summary is reasonable, except that I'd point out that the Africa/Eurasia connection in ALSO not a true separation of land masses. Though this remark is an aside and not strictly relevant to this article about Europe, I think that there is no true distinction to be made between "direct" and "indirect" migration if, as in Battaglia's hypothesis, there was a relatively fast movement by hunter gatherers. Anyway so what? What is it in the wiki article that we are actually debating? I think some of this debate is off topic and getting confused. Perhaps this is because the current article focuses too much upon very old work in Cavalli-Sforza. C-S's "classical" work, as it is called in the wiki article, can be mentioned perhaps, but it has basically been surpassed by work on DNA which he has himself been heavily involved in. I don't think anyone disagrees that E-M78 and E-M81 should be mentioned, but the way this is done is currently a mess.--Andrew Lancaster (talk) 05:42, 15 May 2009 (UTC)
- OK I want to straighten something out. I have not read extensively about this, but have read a bit and know something of genetics. Hxseek states that "Y DNA data do not show any significant direct migration from Africa", this is clearly incorrect. I don't think anyone, except a small number of multiregionalists, disputes that our species arose in Africa. And it's clear that the founder of all extant Y chromosome clades leads us back to Africa. So we are all descended from people who directly migrated from Africa. But even if we discount our initial African origins, I am under the impression that many Haplogroup E subclades are thought to have originated in north eastern Africa and subsequently migrated into the Near East and Europe. Hxseek seems to be saying, when he says "Y DNA data do not show any significant direct migration from Africa", that no haplogroup E clades that currently occur in Europe arose in Africa. I'd like to know the source of this claim. ISOGG states that "Y-DNA haplogroup E would appear to have arisen in Northeast Africa based on the concentration and variety of E subclades in that area today. But the fact that Haplogroup E is closely linked with Haplogroup D, which is not found in Africa, leaves open the possibility that E first arose in the Near or Middle East and was subsequently carried into Africa by a back migration." This is not an equal statement, ISOGG is clearly saying that it is more likely that E arose in Africa, but that there is some possibility that it arose int he Near East. But Hxseek also says "As for E M78, from what I know, it originated somewhere in northeastern Africa", which seems to contradict his claim that there was no direct migration. Actually I'm not entirely sure what he means by "direct migration", and I'm not sure it's relevant, the initial map and caption made no claim for direct migration anyway. In some respects I think this is a red herring, isn't the consensus that haplogroup E and it's subclades are generally involved with bi-directional gene flow between south west Asia and north east Africa? And isn't it clear that there is evidence that during the neolithic there was gene flow from the Near East into Europe? I don't think it's OR to say that haplogroup E might indicate some African gene flow into Europe. In fact I don't think there is any dispute, either from historical or genetic evidence, that African genes have flowed into Europe. We know that there are historical contacts between north east Africa and south east Europe, e.g. during Egyptian and Roman times. But what I find most odd about this debate is they way people are making absolutist claims. I don't understand Causteau's initial claims at all. He starts by stating that Cruciani "says nothing about the "spread" of E-M78 much less from where that spread originated", that's fair enough, but that is not a good reason for not including the map, it is a good reason for modifying the language of the caption. Indeed Cruciani may not say that, but it is well understood that the highest variation of a haplogroup in a population is generally considered to be the point of origin, it's not necessarily important what Cruciani does not say, but it's important to include accurately what he does say. So what does the paper say about the origins of this haplogroup? Then he goes on to say "In fact, Cruciani et al. (2007) indicate that E-V13, the most common sub-clade of E-M78 in Europe, originated in situ in the Balkans during the Bronze age.", I doubt that Cruciani says this at all. Cruciani probably says that based on his data set, the Balkans is the most likely place for E-V13 to have arisen. Good scientists very rarely make unhedged statements, science is based on evidence and hypothesis, so they nearly always couch things in terms of evidence supporting or detracting from any given hypothesis. But again, this is not a good reason for not including the E-M78 map. Clearly E-V13 is a sub-clade of E-M78, so it is absolutely clear that the E-M78 map is relevant to the article. If I can be frank, this looks to me like nothing more than an attempt to try and exclude any data that suggests that there has been recent African gene flow into Europe, and if I'm honest I don't know why anyone who is interested in neutrality would want to do this. Both points of view have some validity, but there is no right and wrong here. Hapolgroup E does provide evidence for north east African Y chromosome gene flow into Europe, and E-V13 may well have arisen in Europe. But to pretend that one of these has greater importance than the other is basically a breach of our neutrality policy. As for claims of African/European admixture, I don't think that's relevant to the original inclusion of the map, neither the map, nor the caption made this claim, it simply stated "The spread of haplogroup E-M78 from Northeast Africa", if Causteau has a problem with the word "spread", then that's easily remedied, simply change the caption to something like "Frequencies of haplogroup E-M78 by geographic region" or something like that. My problem with this map is that the intensity of shading should reflect well defined frequencies of the haplogroup, but these are not given, what is the proportion of E-M78 in the intense orange populations compared to the lighter yellow? It is important to include these frequency data in my opinion.
- One final point, there is some discussion of principal components analysis of classical protein polymorphisms. I don't think that's relevant. Indeed that's synthesis. If we are discussing Y chromosomes, then let's sick to that. If anyone here is going to use data from different studies to produce their own theory, then that's OR and a synthesis. If we are discussing a Y chromosome map, and whether it should be included, then discuss what the authors of the paper that the map comes from say, and discuss the relative merits or including the map, or excluding it. But any discussion of whether autosomal data support Y chromosom data or not should be left to those who write reliable sources, that we can then cite. It really is OR for us to try and synthesis data from different sources, that use different sampling techniques, and that measure different things, and claim that these support the pov that we prefer. Remember we're here to cite what reliable sources say in a neutral way. Frankly I think there is a certain amount of different people pushing their own favourite theory while rubbishing the theories they don't like. We're supposed to work collaboratively. Don't dismiss a map just because you don't like it, or just because you have a problem with the caption. Why not edit the caption so it doesn't make a claim that the original paper doesn't make? Try not to take absolutist positions, and try to see others' points of view. Alun (talk) 06:27, 15 May 2009 (UTC)
- Thanks for your opinion Alun, but I'm afraid you are missing both my and Hxseek's point. I suspect this has to do with your misinterpretation of what Cruciani et al. (2007) mean when they speak of "Northeastern Africa", which is a fairly common and understandable error. It's like this:
- Cruciani et al. (2007) indicate that E-M78 originated in "Northeastern Africa". However, in their study, Northeastern Africa refers strictly to the Egypt/Libya area (please see Table 1 of that study or Dienekes). It does not refer to the Horn of Africa, which the authors term "Eastern Africa". This naming dichotomy is very consistent throughout Cruciani's other studies as well.
- The recent admixture in Europe from North Africa & the Near East comes in the form of E-M81, not E-M78. This admixture is chiefly concentrated in Iberia (particularly Portugal), and is usually seen as the mark of Berber/Islamic expansions.
- Battaglia et al. (2008) indicate that E-V13, the most common sub-clade of E-M78 in Europe, originated in situ during the Bronze age; it was not brought there recently through Berber/Islamic incursions. Cruciani et al. (2007) suggest that E-V13 may have arisen instead in Western Asia during the Neolithic and then entered Europe. That too is not recent admixture.
- User:Wapondaponda has added a map of E-M78 with a caption that reads: "the spread of haplogroup E-M78 from Northeast Africa". This map is sourced to Cruciani et al. (2007) and claims to be based on that study's Figure 2A. However, Figure 2A of Cruciani et al. (2007) indicates the map is of "the observed haplogroup/paragroup frequencies." It says nothing about the "spread" of E-M78 much less from where that spread originated (no mention of Northeast Africa). In other words, Wapondaponda has added a map showing the distribution of E-M78 in Africa, the Near East and Europe, but has modified its caption to make it seem as though it is, in fact, a map showing the radiation of E-M78 chromosomes from Northeast Africa and outwards (which it isn't). He has then added this map to the section of the Genetic history of Europe article titled "North African/Near Eastern admixture" to create the impression that this widespread distribution of E-M78 in Europe represents recent "admixture" from Africa -- when of course, that distinction goes to E-M81, not E-M78 which is of ancient provenance; this is why the section in question's discussion of E1b1b's sub-clades is restricted to E1b1b1b (E-M81) -- Sub-Saharan admixture no less as he has repeatedly insisted. That is original research, pure and simple, since Cruciani et al. (2007) make no such assertion. Causteau (talk) 11:15, 15 May 2009 (UTC)
- Small remark: Battaglia et al. (2008) only say E-V13 might have originated in the Balkans. In fact their wording implies it is likely to have been in either the Balkans or Southern Turkey. They seem to want to emphasize the Balkan possibility but they do not say it outright, and their data favours Southern Turkey. Furthermore, they only manage saying any of this by totally ignoring Cruciani et al's Druze data, and making illogical remarks about the lack of ancient clades in modern places.--Andrew Lancaster (talk) 11:48, 15 May 2009 (UTC)
- Battaglia et al. (2008) state unambiguously that E-V13 originated in Europe: "In addition, the low frequency and variance associated to I-M423 and E-V13 in Anatolia and the Middle East, support an European Mesolithic origin of these two clades." Causteau (talk) 12:00, 15 May 2009 (UTC)
- Hmm. I think this means the abstract states different conclusions than the "real" conclusions in the main text, which, similar to implying that Cruciani et al. had shown no signs of higher STR variation in the Levant, or to implying that 2 cases of E-M78* in the Balkans proves explains that E-V13* happened in Europe, is problematic. Of course it presents us with a problem as editors trying to find neutral sources. The Battaglia paper definitely needs to be treated very seriously where it is clear, but what do we do when it states things in an inconsistent or apparently wrong way?--Andrew Lancaster (talk) 12:10, 15 May 2009 (UTC)
Personally I think Hxseek's remarks about direct/indirect point at what is confusing the discussion (and the article) and also at how we can find common ground. No one is really arguing that Europe was colonized only by boat (direct in the extreme) from Africa, and no one is really arguing against the consensus idea being that all lineages eventually go back to Africa. The problem is that people want to emphasize different leanings on this. I think the best solution requires re-structuring of the article, breaking the different types of evidence and discussion into different geographical regions or entry directions. In this way we can then group the older work by Cavalli-Sforza with new evidence about similar subjects in a legitimate way.--Andrew Lancaster (talk) 06:48, 15 May 2009 (UTC)
Alun, of course all lineages trace to Africa. But if we are going to construct a good quality article, we need to be precise. I think we need to define the concept of movements, or migrations. Gseneticists belive that they can actually decipher whether a movement is a sudden demographic expansion, as opposed to a slow diffusion (based on STRs, star-clusters, etc). Yes, early studies noted the presence of E-M78 (and J2, for that matter) to be reflective of movements from northern Africa and Middle East, resp, and this is mentioned, thereby adhering to neutrality rules. However, better resolution now enables us to understand that the picture is far more complicated. But to say that V13 represents a migration from northern Africa is misleading, because its expansion within Europe has been linked to entirely different demographic phenomena. Hxseek (talk) 09:00, 15 May 2009 (UTC)
- I agree with Alun that Y-chromosomes and classic protein polymorphisms should remain separate topics. However, what is interesting, though need not be included, is that the results from the blood proteins and principal components are consistent with modern DNA studies. Cavalli-Sforza had proposed, based on the classical polymorphisms that there was significant bidirectional gene flow between Eurasia and Africa. However he could not accurately date when the gene flow occurred and also suggested that gene flow could be the result of multiple migratory events. All what he indicated seems to have been confirmed by recent y-chromosome data, and to a lesser extent mtDNA data. Wapondaponda (talk) 10:01, 15 May 2009 (UTC)
Sykes and Oppenheimer
I don't believe the article is as it should be if it relies on a paragraph like this one in the current version:
- Worldwide DNA studies have shown that a group of Homo sapiens left Africa for the Yemen some 80,000 years ago. Some of their descendants entered Europe about 30,000 years later. The Y-chromosome and the mtDNA haplogroups found in Europe differ in their frequency distributions from those in Africa, Asia and the Middle East. However some predominantly European haplogroups are found in Asia and vice versa. A similar correspondence is found between North Africa and Europe.
Can't we find better sources?--Andrew Lancaster (talk) 20:10, 14 May 2009 (UTC)
- I agree. I'm not inspired by that paragraph Hxseek (talk) 00:29, 15 May 2009 (UTC)
- I think if the article is to be an accurate reflection of what is published it should explain that there are different theories and still not consensus. Do others agree? Sykes and Oppenheimer are popularizers, but often frustratingly silly in the way in which they lead people to believe that so many details are known with certainty.--Andrew Lancaster (talk) 05:30, 15 May 2009 (UTC)
- Actually whether Sykes or Oppenheimer are "silly" is a matter of opinion and not fact. I don't think there is any doubt that both are recognized experts. Indeed Misplaced Pages should be using secondary and tertiary sources whenever possible, rather than primary sources. As for the claims in the quote you give, I don't think they are incorrect, and I don't think you can say that this is a claim only made by Sykes or Oppenheimer. My problem with the quote is simply that it is badly written. All the quote states is that we derive from Africa, that AMH entered Europe about thirty millenia ago, and that the ratios of haplogroups in Europe are different to those in the closest geographical regions. Those claims can be supported by multiple sources. I think the real bone of contention regarding the genetic history of Europe is that regarding the relative contributions between paleolithic Europeans and neolithic Europeans. Y chromosome and mtDNA analysis has indicated a majority paleolithic origin, whereas autosomal analysis has indicated a greater neolithic contribution. So was the neolithic a demic diffusion, or was is a cultural transmission? Most papers concentrate on this debate, and that is what the article should concentrate on as well. There are at least tens of papers that deal with this. Alun (talk) 06:42, 15 May 2009 (UTC)
- The quote also makes specific remarks about a Yemen route as if they are uncontroversial. The remarks are out of date and do not represent a consensus amongst experts. Putting them in without further comment is misleading.--Andrew Lancaster (talk) 07:44, 15 May 2009 (UTC)
I also should point out in case it is not obvious that this is a fast moving field, and so it is difficult to find complete up-to-date "tertiary" sources in the sense you mean. In any case these two sources now unjustly dominate parts of the article, and are quite simply out of date, and not a fair representation of the state of affairs. This needs to be changed, because the impression given now is misleading and confusing. Out of date material can be mentioned if it seems important to explain the history of an idea, but not if it is included in a misleading way.--Andrew Lancaster (talk) 08:04, 15 May 2009 (UTC)
Sorry, but there is more to be said: extended discussions about Clan Jasmine and Milesians from Spain are not tertiary material in the sense intended above. They are pure speculative material which does not come from the genetics literature itself. When a NASA scientist writes about UFO's in a paperback, this does not make UFO's scientifically proven - not even if the book sells well.--Andrew Lancaster (talk) 08:14, 15 May 2009 (UTC)
Yep. I agree with Andrew. It's the Yemen bit. I've never heard about that supposed route. The heavy use of Oppenheimer and Sykes skews the article to be to 'British-centred', although I acknowledge that they are prominent figures. Also, Alun, I would not agree with your statement that autosomal data supports a Neolithic origin. Yes, autosomal data do, in general, show a NW - SE trend, but from what I have read, it is only Chikhi that actually links this as evidence for a major demographic expansion of farmers from M.E. Hxseek (talk) 09:06, 15 May 2009 (UTC)
- Just as an aside, this NW-SW trend seems to be to be one of the major trends (clines etc whatever word you like) that have consistently turned up in every type of genetic study since Cavalli-Sforza. These are the raw data which need to be presented, but in many cases the timing and causation of these patterns are still a matter for debate and speculation. This is why I think it is a bit problematic to divide this article up in terms of periods, and why I am thinking using Cavalli-Sforza's 5 basic principal component "trends" might be a neat solution?? This keeps the article neutral because we do not have to discuss genetic clines under categories of periods which already imply that we take a side concerning theories about what caused the clines.--Andrew Lancaster (talk) 11:56, 15 May 2009 (UTC)
The E-M78 map
There is a problem with this map. It has been put into a section which is discussing migrations which are thought to have happened in relatively recent "historical" periods, and specifically it seems that E-M78 (E1b1b1a) is being confused with E-M81 (E1b1b1b) from North Africa.--Andrew Lancaster (talk) 07:46, 15 May 2009 (UTC)
Haplotype V (p49/TaqI)
The paragraph about this subject should be removed. Whoever included it apparently does not realize that this haplogroup designation was long ago replaced by E-M35, and comments about it showing North African migrations have been improved by the discovery of the E-M35 sub-clade, E-M81, which is already discussed in the article. Furthermore, there is much more data since the days of "Haplotype V (p49/TaqI)". The current way in which the article seems to treat these as separate haplogroups is in any case simply wrong.--Andrew Lancaster (talk) 07:49, 15 May 2009 (UTC)
heading: "historic non-European admixture"
This section heading gives a misleading and strange effect. When we speak about the components making up European genetic diversity, we are always talking about "admixture" from outside Europe, so why use this term only for a section about Berbers in more recent millenia? On the other hand it would also be strange to mention "non-European admixture" at every heading. The emphasis of the section titles should only be on the period, OR else the geographical vector of entry being discussed. In fact, I believe the article should be structured around vectors of entry instead of periods by the way, because for each case of clear genetic immigration there are debates about which period it happened in, including E-M81. This would also help make the Cavalli-Sforza "classical" work relevant, because their principal components fit well within later work when looked at this way.--Andrew Lancaster (talk) 07:58, 15 May 2009 (UTC)
Section: Haplogroups in Europe
This breakdown of modern European Y and mt haplogroups is redundant. It is only relevant if the haplogroups are explained in terms of their historical affect on European genetic diversity, and indeed they are where necessary. These remarks should be integrated into other parts of the article or else dropped as something better discussed at Demographics of Europe or perhaps a new article about European Genetic Diversity for example.--Andrew Lancaster (talk) 08:19, 15 May 2009 (UTC)
For now I have left this section in. But perhaps it should eventually be split into a new article?--Andrew Lancaster (talk) 11:50, 15 May 2009 (UTC)
sub-sections: Western Europe substructure and Eastern Europe substructure
Whatever the original intention, these sections are apparently now entirely focused upon the concept of LGM refugia. According to the logical structure of the existing article, discussion about this entire concept, without undue focus upon Iberia and Ireland, should be in the sub-section called Paleolithic and Mesolithic migrations. It should not be separated out and treated twice. I would propose that this section in turn should be divided into pre-LGM, and post LGM sections.--Andrew Lancaster (talk) 08:24, 15 May 2009 (UTC)
Chronological sections have been tidied up now and made consistent, but I still wonder if a chronological approach is best. In any case, whichever way the material is divided up for discussion the article needs one coherent structure.--Andrew Lancaster (talk) 11:52, 15 May 2009 (UTC)
Upgrade, cont
Since we all seem to be in the mood for some changes, I will cont from above. I suggest that we include something about genetic studies, so the average reader can understand them more, their limitations and assumptions. This will hence put the rest of the article in a better context, in that nothing is definite. A discussion on European sub-structure, without the ad nauseaum discussion of R1b prevelance in western Europe. Then Progress chronologically about European mirgations, as theorized by genetic data, incl. the so-called 'classic' stuff incoporated into it.
Here is what I have done so far ...
- ===Genetic studies===
- One of the first scholars to perform genetic studies was Luigi Luca Cavalli-Sforza. He used ‘classical genetic markers’, which are an indirect way of analysing DNA. This method studies differences in the frequencies of particular allelic traits, namely polymorphisms from proteins found within human blood (such as the ABO blood groups, Rhesus blood antigens, HLA loci, immunoglobulins, G-6-P-D isoenzymes, amongst others). Subsequently his team calculated genetic distances between populations, based on the principle that two populations that share similar frequencies of a trait are more closely related than populations that have more divergent frequencies of the trait. From this, he constructed phylogenetic trees which showed genetic distances diagrammatically. In addition, they performed principal component analyses on the data, from which they generated ‘synthetic maps’. These maps revealed several clinal patterns. Genetic clines can be generated in several ways: including adaptation to environment (natural selection), continuous gene flow between two initially different populations, or a demographic expansion into a scarcely populated environment with little initial admixture with pre-existing populations (Archaeological Genetics; The peopling of Europe, page 390). Cavalli-Sforza connected these gradients with postulated pre-historic population movements based on known archaeological and linguistic theories. Given that the time depths of such patterns are not known, “associating them with particular demographic events is usually speculative” (Rosser 2000).
- Studies using direct DNA evidence are now abundant, and may be utilize mitochondiral DNA, Y-chromosomal DNA or autosomal DNA. The first two are particularly popular. MtDNA and the non-recombining portion of the Y-chromosome (henceforth NRY) share some similar features which have made them useful in genetic anthropology. These properties include the direct, unaltered inheritence of mtDNA and NRY DNA from mother to offspring, and father to son, respectively, without the ‘scrambling’ effects of genetic recombination. We also presume that these genetic loci are not affected by natural selection, and that the major process responsible for changes in base pairs has been mutation (which can calculated) (European Prehistory. Sarunas Milisauskas, page 58). The smaller effective population size of the NRY and mtDNA enhances the consequences of drift and founder effect relative to the autosomes, making NRY and mtDNA variation a potentially sensitive index of population composition (Semino 2000; Rosser 2000, Richards 1998). However, these biologically plausible assumptions are nevertheless not concrete. For example, Rosser suggests that climactic conditions may affect the fertility of certain lineages (Rosser 2000). Even more problematic, however, is the underlying mutation rate used by the geneticists. They often use different mutation rates, and can therefore arrive at vastly different conclusions (as we will see later) (Rosser 2001). Moroever, NRY and mtDNA may be so susceptible to drift that some ancient patterns may have become obscured over time. Another implicit assumption is that population genealogies are approximated by allele genealogies. Barbujani points out that this only holds if population groups develop from a genetically monomorphic set of founders. However, that the populations who colonized Europe throughout pre-history were polymorphic has been deemed likely by Barbujani (Genetics and the population history of Europe Guido Barbujani* and Giorgio BertorellePNAS January 2, 2001 vol. 98 no. 1 22-25. ). Moreover, scholars assume that, for example, modern Basques and Near Eastern peoples are proxy representatives for Mesolithic Europeans and Neolithic agriculturalists, respectively. This assumption has, however, been questioned by Levy-Coffman (Levy-Coffman 2005).
- Although sharing similarities, there are some observed differences between mitochondrial and NRY DNA. The Y chromosome, which is much larger, is particularly useful because it has many slowly mutating biallelic markers (SNPs) to help resolve genealogical clades as well as rapidly mutating microsatellite markers (STRs) to aid in the dating of very recent events (Y Chromosome Evidence for Anglo-Saxon Mass Migration. Michael E. Weale. . In addition, the NRY and mtDNA haplogroup (Hg henceforth) patterns appear to be different. As it is apparent below, Y haplgroups show a high degree of geographic structuring, whereas mtDNA haplgroups are more uniformly and ubiquitously distributed. This may be due to socio-cultural factors, namely the phenomena of polygyny and patrilocality (Rosser 2000).
- Whereas Y-DNA and mtDNA Hgs represent but a small component of a person’s DNA pool, autosomal DNA has the advantage of containing hundreds and thousands of examinable genetic loci, thus giving a more complete picture of genetic composition (eg see Seldin). However, descent relationships can only to be determined on a statistical basis because autosomal DNA undergoes recombination and is liable to the process of natural selection.
- Genetic studies operate on numerous assumptions and suffer from usual methodological limitations such as selection bias and confounding. Furthermore, no matter how accurate the methodology, conclusions derived from such studies are complied on the basis of how the authors envisages the data fits with established archaeological or linguistic theories. The relationship between language, archaeology and genetics is a precarious one.
- ===European population structure===
- ====Relation to other populations====
- From Cavalli-Sforza’s original works, the genetic data found that all non-African populations are more closely related to each other than to Africans; this supports the hypothesis that all non-Africans descend from a single population originating in Africa. The genetic distance from Africa to Europe (16.6) is shorter than the genetic distance from Africa to East Asia (20.6), and much shorter than that from Africa to Australia (24.7). The simplest explanation for this short genetic distance is that substantial gene exchange has taken place between the nearby continents. Cavalli-Sforza suggests that this admixture took place 30,000 years ago.
- A later study by Bauchet, which utilised ~ 10 thousand autosomal DNA SNPs arrived at similar results. Principal component analysis clearly identified four widely dispersed groupings corresponding to Europe, South Asia, Central Asia, and Africa. PC1 separated Africans from the other populations, PC2 divided Asians from Europeans and Africans, whilst PC3 split Central Asians apart from South Asians (Bauchet).
- ====Internal diversity====
- Geneticists agree that Europe is the most homogeneous continent in the world (Cavalli-Sforza; Bauchet, Lao). However, some patterns are discernable. Cavalli-Sforza’s principal component analyses revealed five major clinal patterns through out Europe. He also constructed a phylogenetic tree based on genetic distances. He identified major ‘outliers’ such as Basques, Lapps, Finns and Icelanders; a result of genetic drift due to their relative isolation. Greeks and Yugoslavs represented a second group of less extreme outliers. He explained that yugoslavs did not cluster with any of the below groups due to high levels of internal diversity, whilst he did not elaborate upon Greece’s position. The remaining samples clustered into five major groups. The ‘Celts’ (Irish and Scots) grouped together, as did south-western Europeans (Italians, Spanish, Portuguese). Norweigians and Swedes formed a ‘Scandinavian’ cluster. A ‘Germanic group’ was itself composed of two sub-clusters – a northern Germanic (Dutch, Danish, English) and central Germanic (German, Belgian, Austrian) group. Eastern Europeans clustered together (Russians, Poles, Hungarians), with the Czechs laying intermediate between the eastern European cluster and the central Gemanic one. The French clustered with the ‘Germanic’ group, but he clarified that there was appreciable heterogeneity between southern and northern France.
- Semino and Rosser independently performed PCA analyses based on NRY data from several European popluations. As noted above, Y haplogroups show high degrees of geographic structuring. Semino’s data showed three distinct clusters. Western Europeans, dominated by the presence of Hg R1b, clustered together (Basques, French, Spanish, northern Italians, German, Dutch); as did eastern Europeans, who are dominated by the presence of R1a and I2 (Polish, Hungarians, Macedonians, Croats, Czechs, Ukrainians). A middle eastern group was characterised by J lineages (Syrians, Lebanese, Turks). Greeks occupied an intermediate position between European and Middle Eastern populations (Rosser 2000). However, later studies looking at genetic diversity on a more micro –regional scale have revealed significant internal heterogeneity existing internally within some countries, cautioning us from assuming that frequencies quoted in pan-continental studies are representative of entire national communities or ethnic groups. This complexity prompts caution in equating similarity in the frequencies of one or more Y chromosomal haplogroups among populations and common descent (Di Giacomo 2003).
- Data from mtDNA haplogroups is more homogeneous and lacks clear geographical patterning. Apart from the outlying Saami, all Europeans are characterized by the predominance of haplogroups H, U and T.
- Studies using autosomal DNA have showed various patterns. Chikhi’s 1998 study, using only 7 loci, reproduced the southeastern – northwestern cline represented by Cavalli-Sforza’s first principlal component. A later study by Seldin (2006) used over five thousand autosomal SNPs and showed that the only significant population structuring followed a north – south distribution. Hungarians, Poles, Germans, Irish and Ukrainians clustered into the ‘northern group’, whilst Italians, Greeks, Portuguese and Spaniards clustered into the ‘southern group’ (Seldin 2006 * However, this study had a large number of participants who were in fact Americans claiming a particular European origin, as per grandparent’s place of birth). Another study by Bauchet (2006) found a major separation to lie within a northern-southeastern axis (in agreement with abovementioned studies), as well a second west to east axis. Basques and Finns were again found to lie relatively separate from other Europeans.
- The latest autosomal DNA study, using the largest number of participants and genetic loci to date (300, 000 SNPs), found that, with the exception of usual isolates such as Basques, Finns and Sardinians, the European population lacked sharp discontinuities (clustering), although there was a discernable south to north gradient. Overall, they found only a low level of genetic differentiation between subpopulations, and differences which did exist were characterized by a strong continent-wide correlation between geographic and genetic distance. In addition, they found that diversity was greatest in southern Europe due a larger effective population size and/or population expansion from southern to northern Europe, as expected (Lao 2008)
- Lao’s study corroborated Rosser’s earlier study which found that geography, more than language, is the major determinant of genetic diversity (Rosser 2000). For example, Hungarians speak an Uralic language, but cluster eastern Europeans rather than other Uralic speakers in Asia, corroborating Renfrew’s theory about the role of ‘’Elite Dominance” in language replacement.
Work in progress
Hxseek, can you remember to sign your posts here? I guess I am thinking the same way as you. I think that working on this article SHOULD make most disagreements disappear, because they appear to be confusion based. I hope it will not get too many people upset that I have started moving chunks around within the article. I have tried to explain the biggest things here on the talk page, and in my opinion most of the changes needed doing. There might be cherished citations that can still find a new place in the article, but not in the form they were in.--Andrew Lancaster (talk) 10:04, 15 May 2009 (UTC)
Hxseek, concerning the opening, and the general structure of the article, might it make sense to define Cavalli-Sforza's original principle components and how he described them? This can then help structure later discussion in the article as updates in attempts to explain and describe the same patterns, which are also found in Y, at and mt DNA?--Andrew Lancaster (talk) 10:28, 15 May 2009 (UTC)
- Sure. As long as we don't think that section is getting too lengthy Hxseek (talk) 00:16, 16 May 2009 (UTC)
- It would probably mean re-structuring the article in order to avoid repetition. With my editing yesterday I was trying to use the structure that was already in the article.. BTW I have not done much at all concerning the Cavalli-Sforza section which essentially opens the discussion, and I am thinking you are working on that.--Andrew Lancaster (talk) 05:37, 16 May 2009 (UTC)
Notes: what if we based the structure on the 5 principal components?
Here are some notes, which I have also pasted on my Sandbox. Sourcing needs to be done and many things need to be expanded, but just playing with this idea brings forward many concepts which are currently not in the article. Please consider...--Andrew Lancaster (talk) 15:50, 15 May 2009 (UTC)
Five patterns of genetic relatedness have been identified since the early "principle components" studies of Cavalli-Sforza:
- The exact causes of each of these is the subject of continuing discussion concerning different regions of Europe, as well as different periods of time...
1. A cline of genes with highest frequencies in the Middle East, declining to lowest levels in northwest Europe
- Cavalli-Sforza originally described this as faithfully reflecting the spread of agriculture in Neolithic times. This has been the general tendency in interpretation of all genes with this pattern.
- However, Cruciani et al. (2007) harvcoltxt error: no target: CITEREFCruciani_et_al.2007 (help) says there were at least four major demographic events which have been envisioned for this geographic area:
- The "post-Last Glacial Maximum expansion (about 20 kya)"
- The "Younger Dryas-Holocene reexpansion (about 12 kya)"
- The "population growth associated with the introduction of agricultural practices (about 8 kya)"
- The "development of Bronze technology (about 5kya)"
- Concerning Y DNA, this cline is associated with E-V13, and some clades of the J haplogroup.
2. A cline of genes with highest frequencies amongst the Finnish and Saami in the extreme north east, declining to lowest frequencies in the south west.
- Cavalli-Sforza associated this with Finno-Ugric languages, and links between Europe and Siberia.
- This has recently received more confirmation in autosomal DNA studies. See http://dienekes.blogspot.com/search/label/Finland
- Concerning Y DNA this cline is associated with the N Haplogroup.
3. A cline of genes with highest frequencies in the area of the lower Don and Volga rivers in southern Russia, declining to lowest frequencies in Iberia, Southern Italy, Greece and the areas inhabited Saami speakers in the extreme north of Scandinavia.
- Cavalli-Sforza associated this with the spread of Indo-European languages, which he links in turn to a "secondary expansion" after the spread of agriculture, associated with animal grazing.
- Concerning Y DNA, this cline corresponds to Haplogroup R-M17 (R1a).
4. A cline of genes with highest frequencies in the Balkans and Southern Italy, declining to lowest levels in Britain and the Basque country.
- Cavalli-Sforza associates this with "the Greek expansion, which reached its peak in historical times around 1000 and 500 BC but which certainly began earlier"
5. A cline of genes with highest frequencies in the Basque country, with lower levels in an unclear pattern beyond the area of Iberia and Southern France.
- In perhaps the most well-known conclusion from Cavalli-Sforza this weakest of the 5 patterns was described as isolated remnants of the pre-Neolithic population of Europe, "who at least partially withstood the expansion of the cultivators". It corresponds roughly to the geographical spread of rhesus negative blood types.
- In particular, the conclusion that the Basques are a genetic isolate has become widely discussed, but also a controversial conclusion.
- Concerning Y DNA there have been attempts to associate this cline with R1b, but these are not universally accepted.
- I agree that the principal components are a more comprehensive approach to the genetic history of Europe. At present the article contains a lot of disconnected information that could be presented better if integrated. Wapondaponda (talk) 17:49, 15 May 2009 (UTC)
If we expanded it out would be able to cover everything in this format? For example the refugia question is basically a theory revolving around 3 and 5, and so on. Perhaps people can try editing the version now on my Sandbox to see what we can come up with as an alternative.--Andrew Lancaster (talk) 20:07, 15 May 2009 (UTC)
-Thanks for writing that up, Andrew. I think, for PC 1, we should not include Cruciani's demographic proposals becuase they were not part of C-F's original discussion, not does Cruciani himself make specific allusion to it. Instead, for example, Rosser and Semino did in their 2000 paper, by equating it with the contrasting clines shown by R1b vs J2. In anyway, i think we should work the PCs into the history, as CF saw it. So in the Neolithic period we talk about PC1, and other Y data. Then for alleged Bonze Age IE migrations, we can talk about PC 3 and R1a. etc Hxseek (talk) 00:14, 16 May 2009 (UTC)
Yes, if we leave the C-S section as something separate that we do not compare to more recent data then we can not easily juxtapose different authors who are basically talking about the same genetic patterns, but for example with respect to Y DNA or whatever. Some of them cite C-S and some do not. But if we restructure the article on geographical terms, different from the current chronological terms, then we could juxtapose related studies. So there is a question of whether we restructure or not.--Andrew Lancaster (talk) 06:43, 16 May 2009 (UTC)
The "European Population Structure" section
This section is currently tagged for neutrality concerns, and indeed it has problems. It is tempting to delete it and try to salvage some snippets if necessary.--Andrew Lancaster (talk) 05:39, 16 May 2009 (UTC)
This reference might help!
With interests related to this article, I have had this review article published: http://www.jogg.info/42/files/Lancaster.pdf I think it contains a lot of good references and summaries that will also allow others to search further. Let's hope it helps improve the quality of knowledge and discussion on this subject.--Andrew Lancaster (talk) 12:16, 16 May 2009 (UTC)
Adams et al. study = mess
In the North African admixture section, the study that found the higher presence of North-West African infuence in Iberia was Adams et al.: "mean North African admixture is 10.6%". It should be noted that, as you can see in this table from the same study, for some unexplained reason they regarded the entire E-M78 haplogroup (including E-V13) as recent North African input in Iberia, instead of mostly Balkanic influence. Just compare the frequencies in Iberian and North-West African populations of E-M78: E-M78 rarely occurs in North West Africa, while in Iberia is as common as E-M81. If E-M78 in Iberia was introduced by recent NWA influence, then the NWA sample should have the same E-M78/E-M81 proportion as the Iberian sample. The study found in Iberia a 4% frequency of E-M78 and they added it to the E-M81 (4%) and other typical NWA markers to reach the 10.6 figure. Without accounting all E-M78 as of recent North African influence (as all other genetics studies done in Iberia do) the result is 6.6%, figure also more in accordance with the rest of genetic studies, (even though E-V12, E-V22, and E-V65, less frequent than E-V13 but also present in Iberia should also be taken into account, as in Cruciani et al.)
That study is a complete mess. Just look at the table again and see how they considered all J2 and I in Iberia as of "Jewish" origin, and it will become very clear that they only wanted to inflate the North African and Jewish numbers (according to the study 20% of male Iberians have Jewish ancestry). Look again at the table and you'll see how J2 in Iberia (8%) is much more prevalent than J(xJ2)(1%), while in the Sephardim sample they have a very similar proportion: J2 is 25% and J(xJ2) is 22%. So, the Inquisition sterilized all Jewish J(xJ2) (actually J1) carriers?. Now look at I, in Iberia almost as prevalent as J2 with a frequency of 6%, while in the Sephardim sample is only 1%. If I and J2 were Jewish infuence in Iberia then there would be a similar proportion to the Sephardim sample, so it's clear that most of the I and J2 in Iberia is obviously of non-Jewish origins (as most of E-M78 is of non-NWA origins), specially if we take into account all the E-M78, G, K, Q, I and J2 carriers that had a big impact in Iberia, like Greeks, Germanic peoples, Slavs, Arabs or Phoenicians, notable mention to the Romans, and of course the Neolithic. According to those geniouses everything that is not Paleolithic in Iberia is either Jewish or North African, even when the proportions obviously don't correspond.
That disregard to do a good analysis is even more obvious when you take into account that members of the same team, just months before that study was published regarded in other paper all the J1 and J2 in Iberia and the Balearic Island as of ...Phoenician.--83.33.135.22 (talk) 14:57, 21 May 2009 (UTC)
I think the Adams article contains very important data, and is well done in many ways. That the interpretations of that data are a bit messy has been mentioned a few times around the internet before. Articles like this are great as sources for raw data, which is quite a legitimate need in this type of article, but we have to be careful about treating the interpretative sections as if they are a consensus, because one article can never be a consensus.--Andrew Lancaster (talk) 19:49, 21 May 2009 (UTC)
- I agree that the supplemental raw data is very valuable, but, how it is possible to consider that study "well done in many ways"? It entirely revolves around using that (good) data to make the interpretations I addressed in my previous post: no Balkanic E-M78 in Iberia, all G, K, Q, I and J2 of Sephardim origin even when Iberian and Sephardim J2/J1/I proportions (as happens with NWAn and Iberian E-M78) are very off, ignoring for some unexplained reason other historical E-M78, G, K, Q, I and J2-carriers settlers/populations in Iberia, choosing Basques as representatives of acient Iberians when other ancient groups different to Basques shared the peninsula, forgetting about the Neolithic... I would not call any research that is based entirely on those premises as exactly "well done" --83.33.135.181 (talk) 16:44, 22 May 2009 (UTC)
E3b, North African or Sub-Saharan African
E3b is found in North Africa, East Africa and Europe. But because it is a sub-sub clade of haplogroup E, it is ultimately of Sub-Saharan African origin. E3b a young clade, relative to haplogroup E and it is the only clade of E that is found outside Africa. All other clades are exclusive to Sub-Saharan Africa. Basically it is all but impossible for haplogroup E to be anything other than of Sub-Saharan African origin. Its presence anywhere outside Africa is ultimately due to gene flow from Africa. Cruciani et al specifically state
Recently, it has been proposed that E3b originated in sub-Saharan Africa and expanded into the Near East and northern Africa at the end of the Pleistocene (Underhill et al. 2001). E3b lineages would have then been introduced from the Near East into southern Europe by immigrant farmers, during the Neolithic expansion (Hammer et al. 1998; Semino et al. 2000; Underhill et al. 2001).
The hypothesis that E or E3b are anything other than of African origin are rapidly moving into the realm of fringe, theories. The Asian origin of Haplogroup E or E3b have been implicitly debunked by the evidence from recent studies. But because on wikipedia, we only report what has been written, we are simply waiting for the formal debunking of the Asian origin hypothesis. Andrew Lancaster's recent publication states this clearly:
- In summary, when we look at the siblings and “cousins”of E-M35 in its “family tree” we see very little evidence of origins to the north of the Sahara at all. The Horn of Africa seems to have had stronger prehistoric links with the regions west of it, towards the Nile and the southern edge of the Sahara, than to the Levant. It appears possible that it was only after E-M35 came into being that the E clade became involved in migrations both to the north, to the south.
Wapondaponda (talk) 05:34, 2 June 2009 (UTC)
Edit War by Wapondaponda
Wapondaponda's attemps to make this article an afrocentrist panflet, as well as in Sub-Saharan DNA admixture in Europe were he has already broke 3RR, have produce an edit war. This can not continue as it is! The Ogre (talk) 15:41, 2 June 2009 (UTC)
- I've asked for a 3rd opinion. The Ogre (talk) 15:57, 2 June 2009 (UTC)
- It seems 3O doesn't aplly due to the number of editors being more than 2... The Ogre (talk) 16:11, 2 June 2009 (UTC)
As you and Causteau are the ones doing the deleting here, and Wapondaponda's section is cited and he has tried to explain it on the talkpage, can you both please explain what you are objecting to.--Andrew Lancaster (talk) 18:58, 3 June 2009 (UTC)
I started working Wapondaponda's disputed section in isolation. In isolation it now looks better in my opinion. But here comes the question: why do we now have this section about a geographical origin for migrations within the chronological section? Later in the article nearly everything which is said here is said again, here, here and here. So there is now redundancy, and the logical structure of the article is up the creek. I have suggested before that we could consider restructuring the whole article in terms of immigration routes, but this has not happened, and I admit it would not be simple. So we need to stick to the chronological pattern for now or else we are going to have enormous redundancy. We can have small skeletal sections on immigration routes like we already did, but they should not cover all detail twice. So my proposal is that we should use whatever we like out of this section and put it into the appropriate other sections which already exist.--Andrew Lancaster (talk) 10:51, 5 June 2009 (UTC)
Concerning smaller aspects of the edit war:
- I looked up the Malyarchuk ref just by clicking the link and I see that the abstract specifically mentions M1 being African, just not West African. So just deleting it from the description of Malyarchuk said seems wrong.
- I don't agree with Wapondaponda's constant deleting of the word apparent. This field is often misunderstood as being more certain than it is.
- Can everyone who has done reverts please check to make sure you have not reverted things like spelling corrections? In general, reverts are not a good way to work.--Andrew Lancaster (talk) 11:02, 5 June 2009 (UTC)
- Yes the structure is the biggest obstacle to this article. I think in the long term Cavalli-Sforza's PCs will be the most appropriate way to structure the article. Wapondaponda (talk) 06:24, 6 June 2009 (UTC)
The section on African admixture breaches WP:UNDUE. The 6 % of Iberians who carry 'direct' African lineages hardly warrant an entire section in the G.H.o.E article, certainly nothing which cannot be accomodated in the postulated migrations section and/ or a seperate article on Spanish people. I am inclined to remove it soon, and if parties object, then it will have to come down to a vote Hxseek (talk) 22:30, 20 June 2009 (UTC)
New Section explicitly about regional connections
I have moved all this stuff to a new section: http://en.wikipedia.org/Genetic_history_of_Europe#Sources_of_genetic_admixture_in_Europe_by_region. When you see it all together now you can see how terribly redundant it has become. Can everyone start pruning? --Andrew Lancaster (talk) 11:24, 5 June 2009 (UTC)
Coffman quote
Is quoted Unfortunately, misinformation about these haplogroups continues to pervade the public and media. Haplogroup E3b is often incorrectly described as “African,” leaving a misimpression regarding the origin and complex history of this haplogroup.
This statement is somewhat ambiguous and contradictory. On one hand the authors writes Although E3b arose in East Africa approximately 25,000 years ago and on the other the author states there is a missimpression regarding the origin. There is no standard method of describing haplogroups in relation to populations that harbor them. So in reality there are no African haplogroups, or European haplogroups or Asian haplogroups. There are haplogroups that are frequent in Africa, Asia, Europe, America or Australia but not elsewhere, and we may colloquially refer to them as African, Asian, European, American or Australian. This applies to all haplogroups not just e3b. We can also objectively refer to haplogroups by their most likely region of origin, in which case e3b is indeed "African". However this is complicated by the fact that haplogroups tend to move around and are continuously evolving. This obviously applies to e3b which is found in Africa, Asia and Europe. From a scientific perspective, the Coffman quote is not really useful because the author does not describe what is incorrect about describing e3b as "African". If the origin is the criteria for describing a haplogroup, it is legitimate to describe e3b as African. In short the author prefers not to consider e3b as african but this a personal opinion or choice in how to describe a haplogroup. It may contain some social commentary but it serves no value from a scientific perspective. Wapondaponda (talk) 22:03, 10 June 2009 (UTC)
I agree. There are two clear and extreme ways to interpret the idea of an African clade: that it is a clade that is today only present there, like A and B Y haplogroups, or else all haplogroups are African. However it may reasonably be said that E1b1b is somewhere in between, and it was in any case amongst the last to leave the room, as per the Underwood and Kivisild quote I keep reminding you of Wapondaponda. By the way you might want to see http://en.wikipedia.org/Talk:Haplogroup_E1b1b_(Y-DNA)/Archive_5#Ellen_Coffman-Levy_.282005.29 which I think confirms that Ellen was not looking for this type of confusion, but rather had specific misunderstandings in mind.--Andrew Lancaster (talk) 19:56, 12 June 2009 (UTC)
Protection
I protected the article for a week after looking at its recent history, but its been suggested to me that that was not necessary. If everyone agrees of course I'll lift the protection. Dougweller (talk) 04:24, 11 June 2009 (UTC)
It might be a good idea to ask interested editors to first propose what text they think would represent a reasonable compromise in the sections being reverted the most.--Andrew Lancaster (talk) 14:13, 13 June 2009 (UTC)
"Tat C"
When this page is unlocked can someone please update the terminology which refers to "tat C". I understand that this is a clade with what is now called Y haplogroup N: http://en.wikipedia.org/Haplogroup_N_(Y-DNA)#N-Tat.--Andrew Lancaster (talk) 14:11, 13 June 2009 (UTC)
Percentages
Please show percentages for your map and please don't exagerate the frequency of E3B in eastern Europe Wapondaponda Sincerely The Count of Monte Cristo (talk) 03:30, 21 June 2009 (UTC)
- The source does not say that so you are saying something that a source says something when it does not right? If it does please tell me the page if I find it in the source I will delete it no more de The Count of Monte Cristo (talk) 03:37, 21 June 2009 (UTC) Wapondaponda
Sources
Why do so many people say things without sources in this article? Sincerely The Count of Monte Cristo (talk) 00:20, 22 June 2009 (UTC)
- I will be parousing through and tidying the article hopefully in the coming week. Not every single thing needs to be sourced. Someone recently made some modifications, and perhaps left out citations. Feel free to outline particular comments which you belive need clarifying or are lacking sources. Hxseek (talk) 12:23, 22 June 2009 (UTC)
- I think it is an important point that we should all have some common sense and patience about sourcing on these articles where there has recently been a lot of reverts and edit wars. I have also found it very difficult to line up all the citations again given the amount of things that have been deleted, re-inserted, etc. If anyone sees a missing citation please first post a in the text as a marker, and if you have some extra time try looking through old versions, or google, to find the missing information. Do not delete too hastily.--Andrew Lancaster (talk) 13:12, 22 June 2009 (UTC)
Well said Hxseek (talk) 00:45, 23 June 2009 (UTC)
Iberian refugium theory.
I added some sourced material from a scientific journal about the Iberian refugium theory if you delete it there may be content dispute. To HXSEEK from Sophian otherwise known as The Count of Monte Cristo (talk) 20:45, 25 June 2009 (UTC)
- I deleted it - the abstract didn't mention the Iberian refugium theory, have you got a quote from the article that does? Dougweller (talk) 21:09, 25 June 2009 (UTC)
- SOPHIAN, (1) please do not make polemic comments on talk pages (2) I have followed the link to try to find the justification for your edits and I do not see them in the abstract or pages accessible to non logged-in users. Please can you provide an exact quotation on which you are basing your edits? --BozMo talk 21:13, 25 June 2009 (UTC)
I've just tried to tidy up the new attempt by Sophian to create a properly sourced statement about R1b and refugia. This material was clearly cut and paste from somewhere and needs improvement, but I decided not to just delete it. Sophian, what are the tables being referred to? And how can readers understand the flow of discussion which suddenly mentions ht15 and ht35? Please do not expect that people will always take time this way if you paste material in such a sloppy way.--Andrew Lancaster (talk) 05:24, 26 June 2009 (UTC)
- I think that whole section is an 'eye sore'. It should be worked into the migrations and/or summarized a bit. Why does it focus on an ad nauseum cataloguing of R1b frequencies ?
- Moreover, it appears that Sophonian is being disruptive at times, although his R1a map is good. Yet he blatantly breached copy right of Dupanloup's map. He needs to be watched because he has a tendency of introducing some bizarre/ fringe/ unsourced/ deceptive theories. All this undermines his credibility
- I am aware that some of my edits need eloboration of references. These will be done within the next 2 days
Hxseek (talk) 05:30, 26 June 2009 (UTC)
- I agree that focusing upon 1 Y haplogroup is a bit weird. I also happen to know that what is being written about R1b and refugia does not match recent thinking, although so much of this progress has been done within the "amateur" genetic genealogy field, and the action has been so fast and furious, that we still await an article we can cite as a snapshot of more recent thinking. I believe Cruciani is presenting one. In summary R1b-M269 probably entered Europe after the Neolithic, from the Middle East. The phylogeny under R1b-M269 was unknown when the present sources were written, and changes everything. I know that for example Mike Hammer has acknowledged in public (but not in writing?) that all the old estimates of R1b being old enough to have been in a European refugia are now no longer tenable. This is of course also a problem over on the R1b article, so we could look there for inspiration, but I think it is also difficult there.--Andrew Lancaster (talk) 05:53, 26 June 2009 (UTC)
Really? Yes, in Anthropological Genetics, 'The Peopling of Europe", it states that R1b is much younger than origianally proposed. That throws the Basque being some special representatives of "Old Europe" theory right out the window ! (which i was always suspect of). Barbujani has always maintainted that, in general, we are overestimating the ages of haplogroups . Maybe we are Neolithic newcomers, afterall ! The problem is, I have not seen any recent articles on R1b Hxseek (talk) 06:21, 26 June 2009 (UTC)
Exactly. For example for Hammer's remark, I saw it on several twitters coming from the FT DNA conference in Texas this year. I really look forward to seeing what Cruciani has written but I fear it will be out of date before it gets into the public. It seems he'll be writing about the two major sub-clades we now know of, R1b-U106/S21 and R1b-L21, but the latest news is all the phylogenetic structure that has been discovered between these two clades, and M269, some of which sub-clades are apparently only found in Western Asia.--Andrew Lancaster (talk) 06:48, 26 June 2009 (UTC)
R1a1
I added mention of the fact that there is a sizable presence of R1a1 in Scandinavia, and a small presence in the British Isles. User SOPHIAN added the fact that there was also a sizable presence in Paris and attached a footnote, but I see nothing in the footnote which substantiates the statement. MarmadukePercy (talk) 19:40, 26 June 2009 (UTC) I am not trying to argue I am just trying to list all the parts of Western Europe that have decent frequencies of R1A. The frequency of R1A in Paris is 15.2% (Similar to Scandinavia) go to page 43 of my link to see this. Sincerely The Count of Monte Cristo (talk) 23:19, 26 June 2009 (UTC)
Sorry, but I cannot find any mention on page 43 of the link for R1a1 in Paris. MarmadukePercy (talk) 23:44, 26 June 2009 (UTC)
Look at the table at the very bottom of the page (Note:The word Paris is a Image in that article so you can not find it by using find and search) I don't know why they did it like that but oh well :) Sincerely The Count of Monte Cristo (talk) 23:56, 26 June 2009 (UTC)
- Whilst that's all very interesting, I do not see why we have to include the frequency of R1a1 in Paris ? This is about the genetic history of Europe. Logic dictates that we should give a panoramic view into the overall genetic landscape of Europe, pointing out obvious patterns, not getting bogged down with frequencies of R1a1 in Paris, or how many Galicians sport sub-Saharan DNA ? Hxseek (talk) 02:18, 27 June 2009 (UTC)
- I agree with Hxseek. Let's try to keep our eyes on the forest, and not the trees. MarmadukePercy (talk) 02:48, 27 June 2009 (UTC)
R1 origins
Well article (2001), page 5 re: M173: This finding, as well as geography, strongly suggests that the source of both of these migrations was an ancient Central Asian population.
'Putative' means 'possible', does it not. Secondly, this is the general concensus, although a fewer papers have proposed a southern Asian (ie Indian) origin. Thirdly, where abouts in Asia isn;t even the main point of the discussion in the article. It mainly is intended to highlight the difference between mtDNA which appears to mostly have appeared from the Near East, where as a far more significant proportion of Y Hgs seem to have arisen in more distant parts of Asia- whether it is 'central' asia or 'southern Asia' Hxseek (talk) 03:02, 27 June 2009 (UTC)
- Are you saying that there is a consensus that R1 did not originate in the Near East?--Andrew Lancaster (talk) 09:19, 27 June 2009 (UTC)
I cannot recall any author placing it in the Near East Hxseek (talk) 15:34, 27 June 2009 (UTC)
E3b-North African vs Middle Eastern vs Sub-Saharan African
This issue needs to be clarified. Firstly North African populations as we know them today are not representative of mesolithic North Africans. This is clearly illustrated in the study The Levant versus the Horn of Africa: Evidence for Bidirectional Corridors of Human Migrations. According to this study the majority of Y haplotypes in Egypt, about 59% are of Eurasian origin. 41% of the haplotypes are African of which 32% is E3b(XE3b*) and 9% is E(XE3b), E3b*, A and B. Modern Egyptians are thus an admixed population 59% Eurasian and 41% African based on the Y chromosome.
The Eurasian component having entered Egypt after the Neolithic revolution according to A Predominantly Neolithic Origin for Y-Chromosomal DNA Variation in North Africa. Prior the arrival of Eurasian haplotypes, the inhabitants of Egypt would have been allied with Nilotic populations to the south. It is these peoples who migrated into the middle east during the mesolithic carrying with them E-M78, which would later disperse into Europe. To summarize, African haplotypes entered Eurasia before Eurasian haplotypes entered Africa, and thus contemporary North Africans are not representative of the African migrants who carried E-M78 into Eurasia. In this case the presence of E-V13 in Europe should not be considered "North African admixture" in the sense that it does not represent contact between Eurasians and populations similar to contemporary North Africans. Rather it represents contact between Eurasians and mesolithic African population with Sub-Saharan affinities.
This is no longer an "Afrocentrist" position but is supported by multiple and independent lines of evidence. The three lines of evidence include the classical genetics, E3b itself and archaeological evidence.
Classical studies
"The classical genetics studies" by Cavalli-Sforza were published between 1991 and 1993 before the discovery of most Y-chromosomal haplotypes. Haplogroup DE for example, was discovered in 1994. This particular study was careful to use aboriginal peoples to minimize the effects of recent admixture. So for Africans, Zaire pygmies, Central African pygmies and Senegalese Mandenka were used. North Africans, East Africans and Horn Africans who are likely to have Eurasian admixture, were not sampled. Yet these studies were able to detect some form of pygmoid African admixture in the European population. So this admixture is clearly Sub-Saharan, not North African. Cavalli-Sforza states that the spread of farming from the middle east may have been responsible for this apparent African admixture. ,,. This admimxture has been detected accross several genetic measures. Cavalli-Sforza states:
"There is one important exception to the rule in Table 1, namely that in the first column of the matrix Europe shows a shorter distance from Africa than do all the other continents. The difference is statistically significant and is consistently found with all markers, ranging from “classical” ones based on gene products (blood groups and protein polymorphisms) to DNA markers such as restriction polymorphisms and microsatellites" .
Xavier Bolton (talk) 20:06, 28 June 2009 (UTC)
Identification of E3b
The identification of E3b, which was once thought to be of Near Eastern origin is now known to be of Sub-Saharan African origin.
See the edits I have recently made to the Sub Saharan genetic mixture article. The points you raise above are all linked to the question of "what is that article actually about?" Rather than fighting to delete the article I have inserted an introductory discussion mentioning the problems there are in having a clear definition. Clearly different people will be defining things different ways. For example is E1b1b considered to have left Africa to long ago in order to consider it worth mentioning as an African clade? There is no correct answer.--Andrew Lancaster (talk) 07:47, 29 June 2009 (UTC)
Archaeological evidence
Archaeological evidence-Regarding the African affinities of skeletal remains from Turkey, Ricaut et al state
"From the Mesolithic to the early Neolithic period different lines of evidence support an out-of-Africa Mesolithic migration to the Levant by northeastern African groups that had biological affinities with sub-Saharan populations. From a genetic point of view, several recent genetic studies have shown that Saharan genetic lineages affiliated with the Y-chromosome PN2 clade have spread through Egypt into the Near East, the Mediterranean area, and, for some lineages, as far north as Turkey , probably during several dispersal episodes since the Mesolithic. This finding is in agreement with morphological data that suggest that populations with sub-Saharan morphological elements were present in northeastern Africa, from the Paleolithic to at least the early Holocene, and diffused northward to the Levant and Anatolia beginning in the Mesolithic."Discrete Traits in a Byzantine Population and Eastern Mediterranean Population Movements
Another study of Natufian crania by Loring Brace also detected biological affinities with peoples of Sub-Saharan Africa. The Neolithic revolution is believed to have arisen from the Natufian culture in Israel. Agriculture is thought to have spread by demic diffusion from the middle east to Western and Northern Europe. Brace et al. state
"If the Late Pleistocene Natufian sample from Israel is the source from which that Neolithic spread was derived, then there was clearly a Sub-Saharan African element present of almost equal importance as the Late Prehistoric Eurasian element. The interbreeding of the incoming Neolithic people with the in situ foragers diluted the Sub-Saharan traces that may have come with the Neolithic spread so that no discoverable element of that remained. This picture of a mixture between the incoming farmers and the in situ foragers had originally been supported by the archaeological record alone, but this view is now reinforced by the analysis of the skeletal morphology of the people of those areas where prehistoric and recent remains can be metrically compared.The questionable contribution of the Neolithic and the Bronze Age to European craniofacial form
Xavier Bolton (talk) 19:51, 28 June 2009 (UTC)
Nothing wrong with admixture
All human populations are the result of some form of genetic admixture. Being a single species, whenever human populations have come in contact, they have interbred. This is especially true for hunter gatherers as their populations were low, finding a mate was more of a challenge. Hunter gatherers couldn't afford to be too choosy and are more inclined to overlook language, ethnic or religious differences when looking for a mate. Strict endogamy is more of a recent phenomenon that is associated with post neolithic stratified societies. But even with strict endogamy, just a few exogamous liasons are sufficient to introduce new genes and over a sustained period alter the genetic profile of a population. This has been the case with Ashkenazi Jews who are now genetically similar to European populations.
So their is nothing wrong with Europeans having African admixture or Africans having European admixture. Its simply a natural phenomenon. Some people feel that having African admixture is a terrible thing that they are trying to de-Africanize what is clearly African DNA. This is unscientific and it shouldn't be tolerated. Xavier Bolton (talk) 19:52, 28 June 2009 (UTC)
- Good points. We are still going through the article, and making revisions. So far , I am just up to the Neolithic section.
My issue with C-V's work is that he did not include Nth Africans and Western Asians in his genetic distance calculations, but only Europeans, far east Asians and sub-saharan Africans. Although he gave reasons as to why he did not, I think they would clarify the real genetic relationships between Europeans and the rest.
The origins of V-13 have still not been elucidated clearly. Eg Battaglia sees that it did originate in Nth Africa and had a swift early Holocene spread into Europe, and subsequently spread widely from the Balkans at a later period. However, we need to be careful with associating Y haplogroups with ethnicity/ race. Not only is this scientifially incorrect (Y-DNA does not carry 'racial traits'), but it might offend some people. It is nice that you such a positive outlook, but not everyone shares your opinions. Afterall, the NRY is a kind of genetic vestige. It tells nothing about the actual genetic composition of modern populations. Therfore, instead of labelling E3b as "African", one might better state that the mutation defining the said haplgroup first appeared in Africa, or whatever the case may be.
As for jews, they have clear 'Middle Eastern lineages'
Hxseek (talk) 03:10, 29 June 2009 (UTC)
- I think Cavalli-Sforza has been clear in stating that North Africans are intermediate between Sub-Saharan Africans and Europe, though more similar to Europeans than Sub-Saharans. He estimates Ethiopians are 60% Sub-Saharan and 40% Eurasian.
- This is true for North-Eastern Africans but obviously wrong for North-Western Africans. At least we should not consider "North Africans" as a whole. And concerning NorthWest Africa, we should distinguish, at least, Saharan Berbers and Mediterraneans Berbers . According to the most recent autosomal by Jun Z. Li 2009 Saharan Berbers (Mozabites) do have Sub-Saharan ancestry but only around 15-20% (http://1.bp.blogspot.com/_Ish7688voT0/R79MXyHURCI/AAAAAAAAAFg/8MeKImXcV34/s1600-h/structurescience.jpg), so it is very likely that Mediterranean Berbers would show very little sub-saharan ancestry in this kind of study. By the way this is strongly suggested by mtDna studies that found sub-saharan L hg at frequencies around 3%-15% in Mediterrean Berbers (less or same level found in South-Portugal) and up to 20-45% in Saharan Berbers. By the way I share my (portuguese) genetic profile with some mediterranean North-Africans (Morocco, Libya, Algeria and Egypt) at 23andme and I even show slightly more sub-saharan African dna (1%) than them (<1%) in the AncestryPainting--DosReis77 (talk) 08:52, 12 July 2009 (UTC)
- I agree that we need to be careful when associating any haplogroup with race, especially since the y-chromosome has relatively few genes. It's major function seems to make a person male but little else. According to Adam's Curse by Bryan Sykes the Y-chromosome is shrinking because of lack of function. Unfortunately there is a tendency, especially in the blogosphere, to associate these haplogroups with issues of race or ethnic pride.
- I think Cavalli-Sforza has been clear in stating that North Africans are intermediate between Sub-Saharan Africans and Europe, though more similar to Europeans than Sub-Saharans. He estimates Ethiopians are 60% Sub-Saharan and 40% Eurasian.
- However, V-13 is a subclade of M78 which according to Battaglia et al may have dispersed from Wadi Howar in Sudan near the border with Egypt. M78 is subclade of E-m35 which originated in East Africa. So it clearly represents a migration of peoples from East Africa to Europe. What is most telling about the "Africanness" of E-V13 is that Cavalli-Sforza was able to detect Sub-Saharan admixture in Europeans using blood proteins. The blood proteins represent phenotype, which means African DNA is found not only in the y-chromosome but in other parts of the genome. It also means that African admixture is being expressed in the actual genes and is not simply part of junk DNA. This analysis was done primarily using the pygmies of Zaire and Central Africa.
- Thus the Mesolithic North Africans from which E-v13 arose would have been similar to other African populations. The North Africans who lived at that time E-m78 came into being were not similar to modern North Africans who are the product of recent admixture between Eurasians and Africans. In Egypt the current Eurasian component is at least 60% NRY whereas in Tunisia the Eurasian component is 70% via mtdna. So when E-M78 is described as being North African, there maybe a wrong impression about the people who lived there 12,000 years ago before the arrival of Eurasians during the Neolithic. Xavier Bolton (talk) 04:50, 29 June 2009 (UTC)
- I think you are both wrong about Battaglia et al. They felt that E-V13 was likely to have originated somewhere in the area of their study, most likely in the Balkans itself. Personally I think they ignored some important information and they should have stuck to Cruciani et al's proposal that it was probably in the Levant, which I still consider the standard. Of course this lineage is said by both authors to go back to Egypt, and in fact arguing about where a mutation happened is often pointless. --Andrew Lancaster (talk) 06:20, 29 June 2009 (UTC)
- I think you have to be careful about equating a Y lineage's movements with a migration. Better to look at autosomal DNA for this.--Andrew Lancaster (talk) 06:20, 29 June 2009 (UTC)
- I think Xavier is not correct about Afrocentrism. Where science comes to a conclusion of African origins, like with E1b1b, then fine. But a strange form of Afrocentrism exists whereby the same African origins position is taken for unscientific reasons for all haplogroups that MIGHT have originated in Africa, even if scientific discussion is inconclusive or even tending away from this. Take for example Y haplogroup DE and mitochondrial haplogroup M. To look at Y lineages, please consider how there are discussions on the internet calling me a "white cultist" because of the fact that I said in my review article that E-M123 originated in the Middle East or Egypt, instead of saying Africa or Ethiopia. Then tell me there is no "Afrocentrism"?--Andrew Lancaster (talk) 06:20, 29 June 2009 (UTC)
Yes. My mistake. Battaglia argues V-13 arose somewhere in southern Balkans/ Anatolia Hxseek (talk) 06:45, 29 June 2009 (UTC)
- There are extreme positions on the Afrocentrism side too, if they are unscientific then they should have no place on wikipedia. It was initially thought that E-M35 was of Near Eastern origin. In the same way that some scholars still refuse to consider Afro-asiatic as being of East African origin despite this being the most parsimonious solution. So there may be a tendency to over-compensate and be suspicious of any hypothesis that does not consider the African origins of shared haplogtypes. Xavier Bolton (talk) 07:09, 29 June 2009 (UTC)
- Affirmative action has no place in science I'm afraid. Proper scientific debate and information should be nurtured, and then the truth will have a tendency to appear. People writing based on arguments they think others will make are a BIG PROBLEM. The worst POV editing I see is coming from two sides with good intentions. If everyone would take the moral high ground, we would all be better off for it, and any remaining POV pictures would look far less justified!--Andrew Lancaster (talk) 07:44, 29 June 2009 (UTC)
- Do you have a published source for saying E-M35 was thought to have Near Eastern origins ever? I know that some casual ideas existed, but did anyone ever argue such a case anywhere? I think that when E-M35 was first distinguished, web masters just pasted in whatever people used to think about E and DE more generally? I have been asking around about this for quite a long time now. It seems to be like an "urban myth".--Andrew Lancaster (talk) 07:44, 29 June 2009 (UTC)
- Concerning Afro-asiatic, I think that you have to distinguish the real authors on this subject from the general public. The main reason I think a minority propose a non African origin is to do with the paradigm that major modern languages were all spread by the first farmers. So for them it seems parsimonious. Most of these people are archaeologists, and therefore more influenced by clear archaeological evidence than linguistic knowledge. Basically there is only one major linguist who proposes a Near Eastern origin, Militarev, and he proposes it right on the border with Africa so to speak. This theory also is affected by another paradigm which has nothing to do with what worries all the POV pushers, and this is that Militarev is looking for ways to link languages into a super family.--Andrew Lancaster (talk) 07:44, 29 June 2009 (UTC)
Genetic drift, founder effect and population bottleneck
I'm fed up with those with various axes to grind (left, right, up, down, inside and out, rascist and politically correct) trying to draw massive conclusions from just a few pieces of selected data. A link to the articles on genetic drift, founder effect and population bottleneck - which are absolute musts for anybody interested in these sorts of studies - will help to at least stem some of this nonsense. Provocateur (talk) 00:53, 6 July 2009 (UTC)
Western Europeans are very different from Eastern Europeans
http://www.plosone.org/article/fetchSingleRepresentation.action?uri=info:doi/10.1371/journal.pone.0005472.s003 Pair-wise Fst between Europeans: Sykes theory is not out of date infact he implys a expansion from Iberia during the neolithic which would be the perfect origin for R1B1B2 (Although it is always possible that R1B1B2 is of paleolithic origin since it is very diffecult to put a time estimate on a haplogroup) R1A and R1B are the 2 largest y-dna haplogroups in Europe so they deserve much attention and your version awards them little atterntion. The Count of Monte Cristo (talk) 17:37, 11 July 2009 (UTC)
- R1b and R1a are already discussed in this article, as is the East-West cline in genetic variation. We should not explain everything two or three times in the same article. (And please remember that R1b is not the only haplogroup in Europe and the East-West differences are not the only differences in Europe.) What other explanations can you give for your edits? These ones do not explain your edits at all.--Andrew Lancaster (talk) 21:03, 11 July 2009 (UTC)
- SOPHIAN, please explain your edits!
- Why does this article need a listing of dozens of R1b percentages, when this is not an R1b article?
- Why does this article need two maps showing R1a percentages?
- Why is R1a in Paris so important?
- Why does this article need to take a side in the argument about where R1b originated, and why are we giving undue weight to one old source?
- And when you paste in your material, and then refuse to allow anyone to change it, can you please try to check to make sure that formatting, and flow of discussion, and spelling etc are all perfect?--Andrew Lancaster (talk) 07:09, 12 July 2009 (UTC)
- SOPHIAN, please explain your edits!
Copyvio
I've just removed some copied from - I've checked the history of the article and the removed bits only show up after the post to the blog. Dougweller (talk) 20:03, 11 July 2009 (UTC)
Please everyone note that this article has had problems over time because of people inserting too much repetitive material, unduly focused on details
Can all editors of this article please consider that this article has had problems over its history coming from people casually adding material that interests them about details (and then sometimes going on to defend it with more effort than they put into the initial edit). This leads to:-
- Extra material being added without first checking to see whether it is already covered, and where and how it should fit into the structure article. This has led in turn to
- repetition,
- broken structure or flow of discussion,
- inconsistent nomenclature and use of terminology not defined in the article (meaning non expert readers will not understand it)
- Frequent problems with casual additions not being sourced or properly sourced, or "polished" enough to keep in various ways.
- Excessive and unbalanced detail being given to very specific subjects, such as R1a in Paris.
Please aim at a high quality Misplaced Pages. Any efforts you expend without taking the above into account may force other editors to delete your work. Do not assume that they will be able or willing to go and find sourcing, fix formatting, etc.--Andrew Lancaster (talk) 09:27, 12 July 2009 (UTC)
Indeed. This is one of those horrible articles in Wiki, where everyone cherry pics information for one purpose or the other. The only two serious books about genetic history in Europe that are out there are those of Stephen Oppenheimer and Bryan Sykes. They deal mainly with the British Isles, but also with Europe, especially Oppenheimer. To see the quality of the article, they are not even mentioned here. This article is of those that give Wiki a bad name. Kun. —Preceding unsigned comment added by 80.30.190.254 (talk) 09:37, 12 July 2009 (UTC)
I do not believe these two sources are what is called for at all! They are out of date and unscientifically written. Please review the talkpage history on this subject.--Andrew Lancaster (talk) 10:14, 12 July 2009 (UTC)
Sub-Saharan DNA admixture
The article Sub-Saharan DNA admixture in Europe was recently deleted. This article was patrolled primarily by Small Victory. Recently Causteau, SOPHIAN and The Ogre were the primary editors to the article. The deletion of the article Sub-Saharan DNA admixture in Europe means that the community rejected the content of the article. The community has rejected the approach that editors to the article were using. Therefore, users should not transfer the same content onto this article. This time, we have administrative recourse, it is no longer a content dispute. If the same content is transferred into this article we can request administrative intervention based on the findings of Misplaced Pages:Articles for deletion/Sub-Saharan DNA admixture in Europe (2nd nomination). Likewise, I will delete the content that has been transferred from the deleted article. Wapondaponda (talk) 13:17, 19 July 2009 (UTC)
- I agree that any use of this material needs to be very carefully agreed to by editors. I really think nothing in that article can be salvaged in a neutral form.
- But I also want to point out once more that this article is currently broken up in one specific way, which is not the only possible way. All material added to this article should be inserted in a way which sticks to the current structure or otherwise we'll have big redundancy problems.--Andrew Lancaster (talk) 13:26, 19 July 2009 (UTC)
It's funny that the person who, just the other day, accused me (falsely) of trying to suppress evidence of Sub-Saharan admixture in Europe is now himself suppressing said evidence as I'm trying to incorporate it into the article (as was originally proposed). Of course, these genetic lineages are present in Europe and must be mentioned in this article that's called "Genetic history of Europe", just like their Central/East Asian counterparts in that same section. You're not seriously going to disagree with that, are you? ---- Small Victory (talk) 13:37, 19 July 2009 (UTC)
- Small Victory, this is clearly a controversial bundle of material you inserted here, and so slowing down seems to be the least we can do. Please first consider two things:
- 1. Please make sure we only put in material which is uncontroversial and neutral and well sourced, AND which is relevant to this article.
- 2. Look at the structure of THIS article and consider where this subject might fit, and indeed whether it is not ALREADY covered. For example this article covers a lot of the haplotypes being considered already. BUT this article is NOT broken into geographical source areas.--Andrew Lancaster (talk) 13:43, 19 July 2009 (UTC)
- The previous approach to the article has been rejected as discussed in the deletion process. Independent editors viewed the previous material as POV fork. So we need a new approach, I suggest not including any material in this article until we reach a consensus. It is even possible that the a separate article can be created, but it has to be neutral. So I agree with Andrew that we need to slow down. At this stage a draft is probably the best way to go. I suggest someone neutral, other than myself or Small Victory, should lay a foundation to the article. Wapondaponda (talk) 14:52, 19 July 2009 (UTC)
Well, here is the "draft" as it currently appears in the article:
Sub-Saharan African mtDNA (haplogroups L1, L2 and L3) and Y-DNA (haplogroups A, B and E (excluding E1b1b), particularly the ubiquitous Bantu marker E1b1a) are present at generally low levels throughout Europe.
Maternal (mtDNA) lineages are the more common and likely represent gene flow from historical events, such as slavery. They've been found in Portugal (6.9%), Spain (2.1%), Slovakia (1%), Italy (0.87%), Finland (0.83%), Bulgaria (0.71%), Bosnia (0.69%), Basques (0.64%), England (0.6%), Greece (0.44%), Switzerland (0.44%), Czech Republic (0.4%), Russia (0.3%), France (0.3%), Poland (0.18%), Germany (0.17%) and Scotland (0.1%).
Paternal (Y-DNA) lineages occur much less frequently. They've been found in Portugal (3%), Albanians from Italy (2.9%), France (2.5%), Germany (2%), Sardinia, Italy (1.6%), Calabria, Italy (1.3%), Austria (0.78%), Italy (0.45%), Spain (0.42%) and Greece (0.27%).
Genome-wide autosomal DNA analyses using the STRUCTURE clustering program, which is designed to accurately detect and quantify admixture, show equally negligible levels of sub-Saharan African admixture in all Europeans, including Iberians.
Please discuss.--Andrew Lancaster (talk) 15:23, 19 July 2009 (UTC)
- I think that putting aside the various statistics pasted in there are only a few short sentences. Basically these sentences are stating a theory. The theory has the following points...
- Certain Y and mt haplogroups are "sub Saharan", and clear well-defined lists of these can be made
- The haplogroups in these lists are not common at all in Europe, and where they appear this is because of slavery
- Mitochondrial types in this sub Saharan classification are more common than Y lineages in this classification in Europe
- (The link, as I understand it is that in some versions of the story being proposed earlier by the same editors on the deleted article it was being argued that slaves who have children are more likely to be women than men.)
- Not one of these proposals can be read in any of the sources cited, or in any other source, unless we draw conclusions based on synthesizing the data ourselves. The sourcing, if I understand correctly, is supposed to work like this (based on extensive discussion over on the now deleted article where all this stuff was cut and paste from)...
- In order to get the near lists of which haplogroups are sub Saharan, comments from genetics papers calling a lineage sub Saharan are cherry picked. This raises the question of whether calling a lineage sub Saharan in one context, means it is always sub Saharan; and the even more controversial point in practice of whether this mean other lineages are known NOT to be sub Saharan. Geneticists simply do not write this way, and it is a gross misunderstanding of the literature. NOTE: There is no source which gives clear well defined lists, and no source which explains in a clear way how you would even define "sub Saharan" in a way which is going to be consistent for every type of discussion. So this is clearly original research.
- Concerning the slavery claim, again there are some sources cherry picked who mention slavery as a POSSIBLE explanation for unexpected looking results here and there in isolated studies. There is absolutely no source stating that slavery is responsible for the lineages designated (in Misplaced Pages) as sub Saharan all over Europe. This is original research.
- There is not even any attempt to source any statement that mitochondrial sub-Saharan lineages are more common in Europe than sub Saharan male lines. This is original research.
- So I see nothing in this proposed section which should be kept. This is the same original research which was recently deleted from another part of Misplaced Pages. It is compressed only.--Andrew Lancaster (talk) 06:23, 20 July 2009 (UTC)
- I think that putting aside the various statistics pasted in there are only a few short sentences. Basically these sentences are stating a theory. The theory has the following points...
- I concur with Andrew. Essentially this is the same POV material that was in the article and the article was deleted because it was a POV fork. Small Victory and Co, rejected my contributions. They also rejected the compromises that Andrew offered. So the deletion process was a referendum on the approach used by Small Victory et al. Andrew has highlighted how the data in the article was used to present a POV that is actually quite unpleasant. Since this approach was rejected by the community, a new approach is warranted. It is only fair, that others who were not given a chance to contribute have their chance to present the material and subject it to the scrutiny of the community. Wapondaponda (talk) 13:06, 20 July 2009 (UTC)
If anyone cares
Regarding the draft, this is what I have picked up. But my experience so far, is nobody cares about verifying content, so some editors are adding content that may be inconsistent with the sources that they add. I will waste some of my energy to illustrate this, because I have some energy left to waste.
Sources not about Europe
The first four references are.
- Martinez et al Admixture estimates for Caracas, Venezuela, based on autosomal, Y-chromosome, and mtDNA markers.
- This has nothing to do with Europe but Venezuela.
- Abu-Amero, 2007, Eurasian and African mitochondrial DNA influences in the Saudi Arabian population, once again, not about Europe but Saudi Arabia
- Richards et al, Extensive Female-Mediated Gene Flow from Sub-Saharan Africa into Near Eastern Arab Populations , Near Eastern, not Europe.
- Goncalvez et al, Y-chromosome lineages in Cabo Verde Islands witness the diverse geographic origin of its first male settlers Cape Verde is off the coast of West Africa, not Europe.
It seems that the editor is trying to find definitions of admixture from sources not related to Europe. This potentially could be original research. Much better to deal with sources that deal directly with Europe and not other places.
Exclusion of E3b from African admixture
The sources cited by footnotes Richards et al, and Goncalvez et al do not exclude E3b from Sub-Saharan Africa. Goncalvez et al state:
E3b, characterized by mutation M35, probably has an east African origin. The group occurs among Ethiopians (Semino et al. 2002) and Sudanese (Underhill et al. 2000) and appears at frequencies 12%–23% in various Jewish populations (Nebel et al. 2001).
and
The total frequency of haplogroups A and E covering 100% of the 276 Guinean Y-chromosomes is 48% in CVN and 46% in CVS. These values represent the maximum proportion of the west African lineages in the islands. However, since E3b has a significantly higher frequency in north Africans and Middle Eastern populations (12%–22% in Jews; Nebel et al. 2001) than in west Africans (6%), it seems likely that the E3b lineages arrived in Cabo Verde largely from a different source.
So the authors acknowledge the African origin of E3b, but assess two possible routes to Cape Verde. Either via West Africa, but most likely via Jewish immigrants from Iberia. In short, we cannot exclude e3b from Sub-Saharan admixture in such a simplistic manner.
I propose including a quote from Shriver et al, 2008, where they state:
We observed patterns of apportionment similar to those described previously using sex and autosomal markers, such as European admixture for African Americans (14.3%) and Mexicans (43.2%), European (65.5%) and East Asian affiliation (27%) for South Asians, and low levels of African admixture (2.8-10.8%) mirroring the distribution of Y E3b haplogroups among various Eurasian populations.
and
In Mediterranean and Middle Eastern populations, African admixture increases proportionately farther south into North Africa and Southeast into Asia. This distribution appeared to mirror that for the ‘‘African’’ Y chromosome E3b haplogroup (Underhill et al., 2001; Jobling et al., 2003; Cruciani et al., 2004)
. Wapondaponda (talk) 05:21, 20 July 2009 (UTC)
Slavery
The is an overly simplistic statement
"Maternal (mtDNA) lineages are the more common and likely represent gene flow from historical events, such as slavery.
This is sourced to Pereira et al 2000 and is somewhat accurately sourced as they argue that the widespread presence of L lineages in Iberia is due to the slave trade. Firstly the authors state
The geographical distributions of both haplogroups were quite different, with U6 being restricted to North Portugal whereas L was widespread all over the country.
So I don't think it is accurate to describe Sub-Saharan admixture as Negligeable. But back to slavery. Pereira et al 2000 state:
The introduction of L sequences in Portugal was tentatively imputed mainly to the modern slave trade that occurred between the 15th and 19th centuries. Both the great number of slaves that entered Portugal and their very diverse African geographic origin are consistent with the data set now reported. However, we cannot exclude some North-African contribution to present-day Portuguese L lineages.
In general many scholars have started to reassess the overly simplistic almost stereotypical assumption that the presence of L lineages anywhere outside of Africa represents the slave trade. A more recent study by Gonzalez et al 2003, actually contradicts Pereira et al's theory of predominantly slavery era admixture. They state.
However, with respect to the sub-Saharan Africa lineages, the recent history of the Black slave trade carried out by the Portuguese (mainly in the 15th and 16th centuries), with a well-documented import in southern Portugal, also be a plausible alternative to explain the presence of these African haplotypes in this region (Pereira et al., 2000). To test this possibility, we compared the proportion of sub-Saharan Africa haplotype matches between the Iberian Peninsula and northwest Africa (0.75%) with those of the Iberian Peninsula and a sample of sub-Saharan Africans from the Gulf of Guinea. The sample includes 45 Bubis from Bioko and 49 individuals from Saˆo Tome´, 32 Yoruba 1, and 72 Equatorial Guineans. The percentage obtained (0.35%) is roughly half of the former, and in addition, the majority of them (97%) are also shared with northwest Africa, although matches between sub-Saharan and North African samples are only 0.95%. These results suggest that, although both prehistoric and historical influences likely contributed to the sub-Saharan African haplotype pool present in the Iberian Peninsula, the former seems to be more important. Our results are in agreement with the gene flow (19.5%) from northwest Africa to the Iberian Peninsula estimated in a recent study of variation in the autosomic CD4 locus (Flores et al., 2000b), and with the evidence of northwest African male input in Iberia calculated at around 20%, using the relative frequency of northwest African Y-chromosome- specific markers in Iberian samples. Furthermore, our results clearly reinforce, extend, and clarify the preliminary clues of an important mtDNA contribution from northwest Africa into the Iberian Peninsula. On the basis of the L1b frequencies detected in Spanish and Portuguese samples (2–3%) and those found in western Africa (10–30%), a significant influence (at least 10%) of North Africans in the Iberian gene pool has also been admitted
Summary, they argue that L in Iberia is actually of prehistoric origin and not from the recent slave trade. This is not reflected in the current draft, or was not reflected in the deleted article. Wapondaponda (talk) 05:21, 20 July 2009 (UTC)
- I agree that the Pereira article is not a valid source for the much stronger claims being put into this article. It does not even claim to be about all of Europe, or all mitochondrial haplogroups. It is about a specific haplogroup, and only in Portugal. But even then it only it is likely that a part of these lineages were due to slave trade. I also agree that looking at the literature more broadly, even this one citation starts to look over-blown and inappropriate.--Andrew Lancaster (talk) 06:00, 20 July 2009 (UTC)
Percentages
Maternal (mtDNA) lineages are the more common and likely represent gene flow from historical events, such as slavery. They've been found in Portugal (6.9%), Spain (2.1%)or (0.36%) , Slovakia (1%), Italy (0.87%), Finland (0.83%), Bulgaria (0.71%), Bosnia (0.69%), Basques (0.64%), England (0.6%), Greece (0.44%), Switzerland (0.44%), Czech Republic (0.4%), Russia (0.3%), France (0.3%), Poland (0.18%), Germany (0.17%) and Scotland (0.1%). Paternal (Y-DNA) lineages occur much less frequently. They've been found in Portugal (3%), Albanians from Italy (2.9%), France (2.5%), Germany (2%), Sardinia, Italy (1.6%), Calabria, Italy (1.3%), Austria (0.78%), Italy (0.45%), Spain (0.42%) and Greece (0.27%).
Andrew refers to such as "Accurate sounding" figures. Wapondaponda (talk) 05:21, 20 July 2009 (UTC)
- There is also no source being given for saying that maternal lineages are more common than paternal ones. Apparently Wikipedians are coming to this conclusion themselves based on looking at particular datasets. This is OR.--Andrew Lancaster (talk) 06:01, 20 July 2009 (UTC)
Structure, Negligeable African admixture
Genome-wide autosomal DNA analyses using the STRUCTURE clustering program, which is designed to accurately detect and quantify admixture, show equally negligible levels of sub-Saharan African admixture in all Europeans, including Iberians
The sources cited include
- Pritchard et al. I could not find in this article, anywhere that states that Sub-Saharan African admixture is negligeable.
- Genetic_history_of_Europe#cite_note-95 states that Auton et al, say the following
Auton et al. 2009 note higher haplotype diversity in Southwestern Europe, and in particular a higher number of haplotypes shared with the Yoruba, for which they propose four possible explanations: 1) West African admixture, 2) North African admixture, 3) the recolonization of Europe from Iberia after the Ice Age, and 4) a combination of two or all three of these, stating that further research needs to be done. However, in Figure S3 A of the supplementary material, which shows the results of a STRUCTURE-based admixture analysis, neither the Spanish nor the Portuguese have any significant membership in the Yoruba (YRI) cluster.
This is a very dangerous case of original research, because this is not what the authors say. They specifically state, and this is a direct quote, so their is no chance of misinterpreting them.
Our analyses also have direct relevance to current debates in human population genetics regarding the extent of historical gene flow among Africa, Europe, and the Middle East Our observation of a north–south gradient in diversity with the highest estimates of diversity in the southern part of the continent is consistent with the initial founding of Europe from the Middle East, the influence of Neolithic farmers within the last 10,000 yr, or migrations south followed by a recolonization of Europe after the last glacial maximum. The unusually high number of haplotypes in South Western Europe is indicative of recurrent gene flow into these regions. Furthermore, when we considered the extent of haplotype sharing with the HapMap YRI population in Europe, we found that the South and South-Western subpopulations showed the highest proportion of shared haplotypes. If gene flow had occurred solely through the Middle East, we would expect the South-Eastern subpopulations to have the highest haplotype diversity and sharing of YRI haplotypes. These two results therefore suggest that while the initial migrations into Europe came via the Middle East, at least some degree of subsequent gene flow has occurred directly from Africa. A potential concern is that the HapMap YRI are not representative of diversity in North Africa, and the levels of haplotype sharing must be interpreted with this in mind. It is currently unclear how patterns of genetic diversity in the Yoruba are representative of the wider region, although genetic similarity appears to decline with distance. Nonetheless, the haplotype sharing between Europe and the YRI are suggestive of gene flow from Africa, albeit from West Africa and not necessarily North Africa. Future studies will hopefully be able to better resolve this question by comparing haplotypes from further populations around the Mediterranean.
My proposal is to include excerpts from this quote, per Misplaced Pages:PROVEIT#cite_note-1, to avoid confusion or even misrepresenting sources. Wapondaponda (talk) 05:21, 20 July 2009 (UTC)
You think that because the 'Sub-Saharan African' article's talk page is gone you can pretend that you weren't proven wrong on these issues? Not a chance.
Re: Sources not about Europe
Those sources don't have to be about Europe because they're not being cited in reference to admixture in Europeans. They're being cited in reference to the haplogroups that they use (and don't use) as evidence Sub-Saharan African admixture. But of course, you already knew that. You just needed an excuse to dismiss them all because you don't like the consensus they arrive at. Too bad the excuse you came up with is beyond lame.
- Hi Small Victory, I had understood that you were using these articles as a source for building up a case. These sources show which haplogroups are to be called "Sub Saharan". Right? And from these definitions, you put together a case using other information from other articles.--Andrew Lancaster (talk) 14:30, 20 July 2009 (UTC)
Re: Shriver et al. 2008
That study is co-authored by one of the founders of DNAPrint Genomics, and it applies that organization's methodology (AIMs), which has been harshly criticized for "involv loci that have undergone strong selection, which makes it unclear whether these markers indicate shared ancestry or parallel selective pressures." That alone bars the study from inclusion in the article.
- Hi Small Victory, that article making the criticism is making a much bigger argument which basically means a lot of what you are citing and arguing should not be cited and included. You should be careful not to cherry a criticism and only apply it to some data you want to delete.--Andrew Lancaster (talk) 14:36, 20 July 2009 (UTC)
On a side note, the authors' E3b explanation for their (already unreliable) findings makes no sense. They found more West African admixture in Iberians than in Greeks, yet Greeks have much more E3b than Iberians. And they totally ignore E3a and L mtDNA, which are actually West African in most cases and much higher in Iberia than in Greece. So maybe, just maybe, they're barking up the wrong tree with their E3b reference, which would be characteristic of the low level of their DNAPrint-style "research".
- Yes, indeed these published authors might have made mistakes. This is something we can argue about any source, which would then lead to Misplaced Pages needing to be a research institution, or else unable to function, and so Misplaced Pages came up with some policies about things like that.--Andrew Lancaster (talk) 14:36, 20 July 2009 (UTC)
Re: Auton et al. 2009
The note I added (as a compromise to you, btw) perfectly summarizes what that study says about the haplotypes shared between Southwestern Europeans and West Africans:
The high number of samples spanning Europe allowed us to investigate geographic patterns of haplotype diversity at a more localized level. We see a North-South gradient in the number of haplotypes present for both H10 and H25 (Figure 3A) with the highest levels of diversity being found in the Southern regions. In particular, South Western Europe has a higher mean number of haplotypes than South Eastern Europe and Western and Central Europe. This is unexpected, as many current models of historical human migration predict numerous migrations into Europe from Africa via the Middle East, and one would therefore expect the highest diversity in the South East, with decreasing diversity moving North and West . The excess haplotype diversity in South Western Europe has at least two possible explanations. First, it may reflect direct migration from North Africa across the Mediterranean. Alternatively, it may represent a recolonization of Europe after a period of glaciation during which the Southern areas of Europe became a refugium for the prehistorical human population .
To address this issue, we investigated the level of haplotype sharing between African and European populations. In the absence of 500K data from North African populations (the HGDP having been genotyped on a different platform), we investigated patterns of haplotype sharing with the HapMap Yoruba (YRI) population. Using the 25 SNP haplotype windows outlined above, we found that South West Europe had the highest proportion of haplotypes that are shared with YRI (Supplementary Table S5). Furthermore, there were significantly more shared haplotypes between South West Europe and YRI relative to South East Europe and YRI (p-value 0.0072; Mann-Whitney U test), which suggests that the unusually high haplotype diversity in South Western Europe is indicative of gene flow from Africa. However, it is perhaps worth noting that this does not preclude the refugium hypothesis from also contributing to the pattern.
A potential concern is that the HapMap YRI are not representative of diversity in North Africa, and the levels of haplotype sharing must be interpreted with this in mind. It is currently unclear how patterns of genetic diversity in the Yoruba are representative of the wider region, although genetic similarity appears to decline with distance . Nonetheless, the haplotype sharing between Europe and the YRI are suggestive of gene flow from Africa, albeit from West Africa and not necessarily North Africa. Future studies will hopefully be able to better resolve this question by comparing haplotypes from further populations around the Mediterranean.
So no, the authors are not at all clear about how to interpret their finding. Whenever they mention gene flow from Africa, they say "suggestive of". Then they propose the alternative explanations of North African admixture and the Iberian postglacial refugium, pointing out that further research needs to be done.
And they don't indicate a direct YRI contribution either. They indicate haplotypes that are shared between YRI and Europeans, which is not the same thing. Open your eyes and look at Figure S3 A in the supplementary material. It shows virtually none of the YRI component in any of the European samples. ---- Small Victory (talk) 13:06, 20 July 2009 (UTC)
- But you admit that the authors do not say as you propose, that gene flow from Africa is negligible, as they suggest gene flow from Africa. You are focusing on their alternate hypothesis, not their main hypothesis. Their main point is that pattern of haplotype diversity is consistent with gene flow from Africa. Their alternate hypothesis is that it is due to the LGM refugium. I have no problem with including their alternate hypothesis. But if you genuinely look at the data, African admixture in Iberia, is consistent with the several independent studies, Pereira et al, Gonzalez et al etc. So the alternate hypothesis is a less likely explanation for the presence of haplogroup L lineages in Iberia. The YRI haplotype sharing is clearly shown in the YRI table in supplementary materials which is about 5%, so they may not even be referring to YRI in this specific case but to haplotype diversity in general.
- Secondly, you highlight their argument that YRI diversity may not be representative of North African admixture and the wider region. This is precisely what I had been discussing. YRI admixture is not necessarily representative of Sub-Saharan admixture. But YRI is always Sub-Saharan. In short YRI admixture will always represent a minimum of Sub-Saharan admixture. The absence of YRI doesn't imply the absence of Sub-Saharan admixture. But the presence of YRI implies the presence of Sub-Saharan admixture. If say, the study were to incorporate admixture from say indigenous East African populations, Central Africans and Nilotic populations. We would expect the proportion of Sub-Saharan admixture to even be greater than YRI. Recall that African populations are genetically very diverse, and YRI is only one population in Africa.
- So I suggest not sidt-tracking from their main argument, and highlighting their alternate hypothesis as their main hypothesis. Rather their main hypothesis should be given prominence Wapondaponda (talk) 13:47, 20 July 2009 (UTC)
- Wapondaponda, I think that these studies look only for similarities and what their words mean is "we do not know which migrations happened we just see these similarities and think they must be caused by something interesting". For example the Yoruba and Iberian might both have a lot of North African admixture. It is the same point I was making on the deleted article talkpage about Ethiopians. When "admixture analysis" clusters Ethiopians as a "mix" between Western Eurasians and "Black Africans" this does not mean the geneticists are saying that there were two source populations who were mixed together to give a cocktail. It means that they pretended something like this in their mathematical analysis, in order to get a perspective. Many different types of migrations can explain a single result like these. Obviously my point applies to Small Victory as well because what I am saying is that any attempt to take one admixture analysis and make big claims about directions of migrations is something we have to be very careful of.--Andrew Lancaster (talk) 14:44, 20 July 2009 (UTC)
- I agree in that, the precise details of prehistoric migrations are not known. The purpose of the Hapmap, was simply to take samples from a "representative population" in each continent, and make their DNA profiles available to the public for study. The hapmap project specifically state:
These samples were collected in a particular community in Ibadan, Nigeria, from individuals who identified themselves as having four Yoruba grandparents. It is important to include a reference to "Ibadan, Nigeria" when describing the source of these samples out of respect for the community's wishes. Including the name of the city and country where these particular Yoruba samples were collected also reinforces the point that the sample set does not necessarily represent all Yoruba people, whose population history is complex. These samples should not be described merely as "African," "Sub-Saharan African," "West African," or "Nigerian," since each of those designators encompasses many populations with many different ancestral geographies. Note that the adjective form is "Yoruba," as in "the Yoruba samples," not "Yoruban." The accent is on the first syllable (YOR-u-ba).
- So in this case YRI is a stand-in for Sub-Saharan Africa, albeit an imperfect one. We can say, with some degree of confidence, that it wasn't Yoruba people who migrated into the Iberian peninsula in pre-Neolithic times, since the expansion of people from West Africa is a fairly recent event, coincident with the Bantu Expansion around 5,000 years ago. However, as I have previously posted, pre-Neolithic North African populations, were probably related to the ancestors of Yorubas and other Sub-Saharan Africans. Over time these North Africans seem to have admixed with incoming peoples from the Near East as hypothesized by Rando et al 1998, and Arredi et al 2004 creating the current genetic structure of Modern day North Africans. The Berbers have a genetic structure of >70% E-M81, which is East African and up to 48% haplogroup L and M1 lineages. Traditionally these L lineages have been ascribed to the slave trade. But this view is changing now, because many of these L lineages are unique to North Africa, per Gonzalez et al 2003. We can therefore state, that the emerging consensus is that the prehistoric North African immigrants to Europe, especially through Iberia, were not contemporary West African peoples, but they were related to them. The Shriver study, basically found the exact same pattern as Auton et al by also using YRI/West African samples. Science is based on replicability. If numerous independent studies converge on the same finding, then we approach a mainstream consensus. Wapondaponda (talk) 15:59, 20 July 2009 (UTC)
- Wapondaponda, I think that these studies look only for similarities and what their words mean is "we do not know which migrations happened we just see these similarities and think they must be caused by something interesting". For example the Yoruba and Iberian might both have a lot of North African admixture. It is the same point I was making on the deleted article talkpage about Ethiopians. When "admixture analysis" clusters Ethiopians as a "mix" between Western Eurasians and "Black Africans" this does not mean the geneticists are saying that there were two source populations who were mixed together to give a cocktail. It means that they pretended something like this in their mathematical analysis, in order to get a perspective. Many different types of migrations can explain a single result like these. Obviously my point applies to Small Victory as well because what I am saying is that any attempt to take one admixture analysis and make big claims about directions of migrations is something we have to be very careful of.--Andrew Lancaster (talk) 14:44, 20 July 2009 (UTC)
- As usual, you have no idea what you're talking about. E-M81 and M1 subclades are indigenous North African markers that arose in populations who had migrated from West Asia. They're no more "East African" than any other haplogroup on earth. And no, Shriver and Auton didn't produce the exact same pattern. Auton's admixture analysis found virtually no Sub-Saharan African admixture anywhere in Europe. Shriver's found elevated levels of admixture that are undoubtedly a product of the faulty methodology used. ---- Small Victory (talk) 13:18, 21 July 2009 (UTC)
Auton et al.'s main hypothesis was given precedence in my note. Out of the four possible interpretations, I listed it first. What more do you want? Should we just ignore the other hypotheses and the results of the admixture analysis? I'll bet you'd like that, but it's not going to happen.
Also, you're still misunderstanding what Auton actually found. Haplotypes shared between the Yoruba and SW Europeans means exactly that, and nothing more. It does not necessarily mean that one population is admixed with the other. They could both be admixed with a third group (such as North Africans, who weren't tested in the study) or it could simply be the result of genetic drift linked to human migrations that ultimately trace back to Africa (hence the Iberian refugium hypothesis).
It isn't up to us to decide which hypothesis will turn out to be correct. All we can do is report what's in the study, which is what I did. ---- Small Victory (talk) 13:01, 21 July 2009 (UTC)
Yes you did mention it, but the text mentioned a lot more of the other hypothesis than the main one.
If two populations share a similar haplotype, then three scenarios apply
- Convergent evolution
- Common ancestry
- Admixture
It is standard for scientists to consider all three scenarios when comparing two populations.
- Convergent evolution does occur but due to the randomness of mutations, it is usually the least likely scenario.
- Common ancestry is also possible because humans are apparently something like 99.97% alike. But for two populations to randomly converge on similar frequencies of rare traits is unlikely. Most likely if two populations converge to have similar frequencies of a haplotype, then they face similar selection pressures.
- Admixture-If there is evidence of contact or gene flow.
Typically the scientists will chose one of these as their main hypothesis by weighing the evidence at hand. So you are right in that haplotype sharing between Yoruba and SW Europe, may indeed be a coincidence. But in this study, they hypothesize in the discussion when the say
Nonetheless, the haplotype sharing between Europe and the YRI are suggestive of gene flow from Africa, albeit from West Africa and not necessarily North Africa.
The acknowledge that they would like to investigate North African populations as well. But my sentiment, is that won't change much. Genetic Diversity is clinal, so most likely there will be a cline from West Africa across North Africa to Iberia of YRI haplotypes. This has already been demonstrated by studies of Berbers which show they occupy an intermediate position between Sub-Saharan Africa and Eurasia. Wapondaponda (talk) 13:40, 21 July 2009 (UTC)
Please Make it easy
Please tell (on this talk page) me how many people where studied to get the 2.1% figure for Spain if you are going to revert my edit The Count of Monte Cristo (talk) 02:21, 20 July 2009 (UTC)
- Most of the mtDNA data comes from Achilli et al., which is a comprehensive survey of Europeans. It includes a large sample of Spaniards from all over the country. Here's the breakdown:
Spain-North-West ... 8/216 ... 3.7%
Spain-North-East ... 3/179 ... 1.68%
Spain-Center ....... 1/148 ... 0.68%
Andalusia .......... 2/114 ... 1.75%
---
TOTAL ............. 14/657 ... 2.1%
- I'll make a general point about this kind of problem, because I have come across it before. If this was a key point for the article, for example if the article was about Iberian DNA, then it might be a good idea to put in more data, but given that this article is about the whole of Europe I think if anything we need less detail. In practice, these are small percentages for these haplogroups and you are talking about the difference between finding 1, 2, or 3 people. If you think the % will mislead people why not write "3 out of 179"?--Andrew Lancaster (talk) 14:26, 20 July 2009 (UTC)
- The 95% Confidence intervals for rows are:
- 8/216 - 3.7%.. 1.9% to 7.3%
- 3/179 - 1.68%. 0.61% to 4.9%
- 1/148 - 0.68%. 0.016% to 3.8%
- 2/114 - 1.75%. 0.054% to 6.3%
- 14/657 - 2.1%. 1.27% to 3.6%
- PB666 22:50, 21 July 2009 (UTC)
Not fair unless you include Basques and data from rhouda et al. The Count of Monte Cristo (talk) 15:55, 20 July 2009 (UTC)
- I have moved the posts so they can be read properly. SOPHIAN, please take more care where you insert words on talkpages. This is not the first time you inserted right in the middle of someone else's posting. People have to be able to read this. Doesn't Basque country come under North East normally? Surely we don't need a statistic for every region in Europe.--Andrew Lancaster (talk) 17:04, 20 July 2009 (UTC)
Rhouda et al. Studied 844 Spanish people and 3 had L (0.355%) Achilli et al. Studied in addition to what you say 156 Basques and 1 had L Leaving 18 cases of L in 1657 Spanish people or 1.09% The Count of Monte Cristo (talk) 21:48, 21 July 2009 (UTC)
Draft
I have started a draft here Talk:Genetic history of Europe/ Sub-Saharan African admixture in Europe. I am just throwing some ideas out and nothing is concrete. But if anyone has some good ideas we can incorporate them into the draft. Wapondaponda (talk) 06:10, 20 July 2009 (UTC)
- I fail to see why the old article had to be deleted only to start over with a new article on the topic. If the content of the old article was flawed, the standard appoach would have been just to fix it. I also fail to see the relevance of the topic as a standalone article. As in many of our genetics articles, we are mostly looking at random detail cobbled together from academic papers directly (as opposed to via a secondary or tertiary, encyclopedic source). This makes for a "better than nothing" collection of factoids, but nothing that can be used to draw any sort of conclusion. Of course there is genetic flow everywhere, even across the Sahara. It is just a matter of "more" vs. "less" admixture. Prehistoric Sub-Saharan admixture in Europe will be practically impossible to measure without (extremely difficult) recourse to ancient DNA because it is so weak that it can hardly be distinguished from Early Modern admixture.
- The result of the AfD is to eliminate the article as it was principally opinion and junk science, and retain a very small bit of the article in the merge to this article, the delete and merge was part of the consensus opinion. I have no great problems with what was written with this regard and I was for the articles deletion.PB666 22:23, 21 July 2009 (UTC)
- In a nutshell, I do not see the point of such an article. --dab (𒁳) 13:20, 21 July 2009 (UTC)
- A more fruitful approach imho would be an attempt to compile a discussion of the major barriers to human gene flow. The Sahara will feature very near the top in that, probably just second to the major Oceans. This would be a topic very relevant indeed to prehistoric human migration and one that I think could be made into a coherent, meaningful article. --dab (𒁳) 13:24, 21 July 2009 (UTC)
- I think recent prehistoric admixture can be measured because it is still present in the European gene pool. Overtime, as the genome gets sliced and diced and people migrate back and forth, it would become impossible to discern. But presently, patterns of prehistoric African admixture are apparent. Wapondaponda (talk) 14:35, 21 July 2009 (UTC)
Editors showing their true colors
Now that we're "sharing space" with another type of admixture in Europeans, we can bear witness to the agenda-driven double standard that's applied by certain editors here (namely Wapondaponda and Andrew Lancaster). Notice that neither one of them has any problem whatsoever with the Central/East Asian admixture section, even though it's constructed identically to the Sub-Saharan African admixture section. They haven't laid so much as a finger on it, and it barely even registers on their radar. There have been no "citation needed" links added to it, no concern expressed about neutrality, no questioning the definition of "East/Central Asian admixture", no attempts to include R1a or R1b (whose ancestors, R and P, originated in Central/East Asia, just like the ancestors of E-M78, E-V13 and E-M81 originated in East Africa), no insistence on qualifiers regarding "older connections" or populations not necessarily being "separate entities", no nitpicky rewrites and deletions of material without cause, and no petitions to have the entire section removed altogether. Only the Sub-Saharan African admixture section is subjected to that kind of scrutiny and that kind of thinly-veiled OR and POV-pushing. I wonder why that is. It couldn't possibly have anything to do with Afrocentrism, could it? Perish the thought. ---- Small Victory (talk) 13:51, 21 July 2009 (UTC)
- I don't think one would consider Andrew an Afrocentrist. Wapondaponda (talk) 13:57, 21 July 2009 (UTC)
- But one would sure consider you an Afrocentrist (which you don't even attempt to deny). And Andrew is turning into your clone. ---- Small Victory (talk) 12:06, 22 July 2009 (UTC)
- I have no idea what the above means, it is argumentative, a rant and should be refactored. Talk pages are for the improvement of the article, they are not a forum, a soapbox, your psychiatrist, etc.PB666 13:59, 21 July 2009 (UTC)
- If you have no idea what it means, then don't make assumptions about what it is. I assure you that it's related to the quality of the article, and its implications are perfectly clear, which is why the response has been so underwhelming. ---- Small Victory (talk) 12:06, 22 July 2009 (UTC)
go it alone editing
Small Victory, concernint this edit, just to remind you again, the deleted article was entirely your baby. You rejected contributions from myself and compromises from Andrew. The community rejected your approach with ]. That was essentially a referendum on your approach. So this time, we have to use another approach. SOPHIAN was blocked yesterday for recreating the article Sub-Saharan DNA admixture in Europe, what you are doing comes awfully close to what SOPHIAN is trying to do. I suggest you accept that you must work with others to achieve a consensus. Wapondaponda (talk) 14:11, 21 July 2009 (UTC)
- You're quite delusional. That article was deleted because it was a WP:CFORK. And your POV-pushing, original research, 3RR violations and sock puppets had more to do with it than anything I ever did. In fact, the article was problem-free until you (and Andrew Lancaster) came along and started tampering with it. Let's remember that you're the one who's been blocked for repeated rule violations. My record is clean. So if anything, the deletion was a referendum on your approach. Take the hint. ---- Small Victory (talk) 12:18, 22 July 2009 (UTC)
Luigi Luca Cavalli-Sforza
Once again I have been invited to come over and see what my fellow wikipedians are up to. Folks, y'all need to put the coffee cups down (or what ever source the caffiene is injected from) and slow down your discussion and remember the talk page guidelines.
Regarding this author - he was a prominent author back in the 60s - 80s. The basic problem is that most of the work came before the age of precise genetic methods. He used serological techniques and a whole slew of methods that are clearly out-of-fashion now. A.C. Wilson and company really introduced the age and genre of information we are now in, and much of ACs conclusions have effectively replaced Cavalli-Sforza's early conclusion, including the OoA and timing of the OoA issues. This article focuses way to much on C-Ss work, it sort of reminds me of the types of discussions that came from books in sci.anthropology.paleo in the mid 1990s. He is still somewhat active with Y chromosome interpretations but alot of the things I have seen him suggest lately are pretty much off by a factor of 50% anyway. I really do not like articles with alot of 'this famous guy' name dropping. Name dropping IMHO is a form of propoganda and excessive use violates WP NPOV guidelines.PB666 14:20, 21 July 2009 (UTC)
- I agree that CS's work is relatively dated, but it was one of the first comprehensive analyses of human genetic diversity. Yes apparently, he only used a few markers to synthesize his principal components. Since the sequencing of the whole genome, it is now possible for more detailed scientific investigations. My impression is that CS's studies have not been completely invalidated by recent studies, rather recent studies have refined much of CS's work. What is great about CS's work, is that it is fairly easy for lay people to understand, which makes it ideal for wikipedia, because wikipedia tends to have a general audience. Of course other's may argue that if it's for lay people, then it is an oversimplification, but we have to live with this dichotomy. Wapondaponda (talk) 14:29, 21 July 2009 (UTC)
- The difference is much like the difference between the Bohr atom and the Heisenberg/Quantum mechanical Atom. Unfortunately one is wrong, the other is not.
- Despite the simplicity of his conclusions, the approach is rather wrong. And if NPOV is to be employed why is Wilson's work not covered. Here is the basic problem, and I apologize if I seem to insult anyone here, this is not my intent. There was a first person who left Africa as such or a group, there is archaeological evidence that this might have occurred 125 kya (both from the levant, but supported by Liujiang). There are archaeological sites in India that date to about 80kya that also support this. These migrations are occurring ealier than the earliest fixation dates for the Y-chromosome, and given the 2N rule we can estimate that a TMRCA for Y chromosome is occurring and constrictive evolution that fixes it for at least 30ky after the TMRCA it places Y-chromosome in a constrictive context in Africa to 90kya which is rougly about the time people are reaching East Asia. While the mtDNA does not suggest migrations this early its confidence interval allows such early migrations. So the discussions about Y-chromosome and timings of migrations as seen in Journey of Man are basically idealizations, not reality. Either there are some mistaken assumptions with molecular clocking of the Y chromosome, or Y-chromosome underwent a post exodus selective sweep, and therefore it is an unreliable marker for early migrations anywhere. Frankly, I think exit dates which include the previous interglacial period (up to 131 kya). In addition, the dates archaeology generally agrees upon is that humans had reached Eastern Europe by 40kya, and that by 34 kya there was occupation within central Europe, by 30 kya they had reached Iberian and were affecting the tool industries of the region (therefore the dates in the article are not correct). There is molecular evidence that there was early geneflow from Africa, and although there is little archeaological support, at least by the epipaleolithic most of the archaeological evidence comes from coastal regions of iberia, suggesting a shift from open range hunting, which could indicate the dominance of one culture during the glacial period which favored eastern migrants but a shift to coastal foraging which favor migrants from the south. There appears to be a meshing of African and Eastern DNA in Iberia.
- Here is my basic problem, migrations and the alleles or haplotypes that these migrations introduce are not general or diffuse, particularly when one talks about crossing bodies of waters. Molecular studies now clearly indicate that many founding or settlement events are quite discrete, both in size and within a temporal context. HLA studies of Iberians and Europeans indicate quite clearly that discrete contributions from Africa have occurred in the past. The problem is not that these discrete events occurred, the problem is how do we create a context which is correct (this was my problem with the Subsaharan page). A recent study of Eastern Islanders for example showed not native American mtDNA or Y chromosomes, but 2 individuals had HLA that came from South America, reproting the fallibility of both analysis. When did this admixture occur, before Europeans or after Easter Island became part of Chile. The result is not unexpected because HLA is under heterozygous selection and mtDNA and Y are haploids with tendencies to undergo fixation. Effectively HLA preserves about 3 times the added diversity of mtDNA and Y combined in real time.
- There is a problem when using mtDNA and Y is that the fix rapidly often leaving the impression that an event occurred once and from a single source(and yet mtDNA will give a date of say 60 kya and Y for the same date 20kya, and the HLA studies will indicate that both dates and other dates might be correct). Scientist compound the problem with what is statistically known as B-error (type-II error), that means not to collect enough data to show a significant separation when a significant separation exists (a major problem I have with C-Ss work). Consequently, the mtDNA people claim there was a single event for exodus, when in-fact, there were multiple events. This is pertinent to European claims because certain mtDNA that appear to have arrived into Iberia are either contemporary to this migration (60 to 130 kya) or different. The archaeology makes no claim that AMHs were in Iberia before 35 kya which leaves 'silent' period of 25 to 70 kya. It is imperative to look at the potential flaws of every claim. So that many discrete events may occur, we often don't have enough data, the proper tools to 'parse' that data objectively, or we don't have a means of properly calibrating the molecular clocks to time the events (For example, these new Mitogenomic clocks have a huge problem with selection at coding sites that corrupts the clock, removal of these sites makes a less precise clock that is more accurate).
- Every claim should be critiqued, the weakest claims, no matter how historic, popular should be pruned from the article. I can give an example of this, the Topper archaeo-philes are presenting now that humans came from Europe to the New World (preclovis) and this is gaining popularity. THere have been claims of sites in S America that Africans came across the atlantic and settled 50 to 60 kya. Even some claims that Homo erectus or Neandertals made it first to the new world. There is no apparent molecular evidence from mtDNA, HLA, or Y supporting these beliefs. In addition these claims are based on tools that are not obviously of human origin.
- Speculative genetic claims, and I know most of them, really cannot be founded in archaeology and the molecular clocking makes some rather large assumptions. It is very difficult to assign migration to a specific paleoanthropological context, and even if one can, the variance on these clockings are -50/+75% about a relative range.
- Equity in speculative claims, if two claims are equally speculative, don't add more speculative claims, instead cite the reasons for why a speculative claim should be removed here, bring in the most recent literature in support of its speculation and remove it.
- Molecular anthropology is what it is. It cannot be dressed up to be made more than it is, the tools we have are frequently abused to create speculation. Many papers from the 80s, 90s and even some recent papers make conclusions that cannot be supported by their data or statistics, many of these have proven to be incorrect. What happened to MREH? What became of Ayala's "Myth about Eve"? The mhc16 study? The recent AfD highlighted the questionable conclusion of an individual who works on HLA, and the situation between Africa and the Greeks. For the mtDNA folks here, be aware the from 2000 to 2004 there were a number of papers published with a fantastice number of sequencing errors, particularly papers dealing with Europeans . From every perspective there are errors in the data set.
- mtDNA has a number of supervariable sites such as 16129 and 16183-16192 that are not useful. 16183-16192 and some other sites have been noted to change from different samples of the same individual. The mtDNA genomic clocking on rarely mutated sites...I simply do not trust, these sites may be variably selective and disappear as soon as people migrate or a culture shift occurs. As stated above there is a rather large history of sequencing errors. Historic comparisons are based on assumptions between HVR mtDNAs and genomic mtDNAs may introduce inaccuracies.
- Y chromosome. It is good to see a refinement of Y chromosomal SNP typing, the STRs had been used in the past and these often betray ancestry. Some older schemes of ancestral typing may not be comparable to the most recent schemes. Beware studies that rely on molecular clocking of Y chromosome, there are assumption errors in the clocking that cannot yet be explained. Y-chromosome may be heavily influenced by cultural selection and fixation could be much more rapid in certain instances that is currently believed. The 2N rules are based on variable selection, if selection is positive for any length of time the 2n rule cannot be applied.
- HLA. HLA suffers from similar historic problems. Early studies typed A9 for instance that then became A23 and A24 and then evolved to A*23 and A*24 which then resolved into A*2301, A*2302, A*2303, ...,A*2402, A*2403, ..... and one has to be very careful when conclusions are made based on old typing technology or typing kits. Cross references need to carefully resolved, for instance why A24 can be called A*2402. There is another problem with HLA, and that is haplotype frequencies are frequently reported as 1 or 2 occurances. The relative frequencies that can give rise that can give rise to 1 to 5 observations have a wide relative variance. This fact also needs to be applied to frequencies based on Y and mtDNA. Let say A paper finds the haplotype A*3108-B*5112 and this is deemed to be of native american origin, how can we be sure unless all A31-B51 that exists in Asia has had the same degree of typing.
- Be critical of research that might be speculative or synthesis based on limited or partial evidence, with comparisons have the proper background work on all possible contributing populations been carefully done. How will the results reflect reality if new mutations in a sequence are found, or if another population it typed with more individuals and thus showing more haplotypes that give it better statistical power.
- PB666 17:54, 21 July 2009 (UTC)
North Africa
This section needs some clarification. North African admixture in this case, should strictly speaking consist of E-M81 and U6. These are the only clades that are specific only to contemporary North African populations. E-M78 has a presence in Sub-Saharan Africa. In addition, and we know that the expansion of E-M35 started in Sub-Saharan Africa, proceeded to North Africa, the levant and then Europe. Currently the initial expansion from Sub-Saharan Africa has been ommitted, and an arbitrary region, North Africa has been selected as the source population. So even E-M81, does also represent the expansion from SSA as well, but its may or may not have involved contemporary North Africans. Wapondaponda (talk) 16:41, 21 July 2009 (UTC)
Pictures of children
A user added a series of photos of children. I don't see what relevance they have in a genetics article, so I have deleted them. Wapondaponda (talk) 18:35, 21 July 2009 (UTC)
Genetics section
I am going to go through the genetics section presenting paragraphs 1 by 1 point out problems with each paragraph.
One of the first scholars to perform genetic studies was Luigi Luca Cavalli-Sforza. He used classical genetic markers to analyse DNA by proxy.
This method studies differences in the frequencies of particular allelic traits, namely polymorphisms from proteins found within human blood (such as the ABO blood groups, Rhesus blood antigens, HLA loci, immunoglobulins, G-6-P-D isoenzymes, amongst others).Subsequently his team calculated genetic distanceSubsequyently the genetic distance between populations was calculated, based on the principle that two populations that share similar frequencies of a trait are more closely related than populations that have more divergent frequencies of the trait. and phylogenetic trees were constructed that showed genetic distances diagrammatically.His team also performed principal component analyses, which is good at analysing multivariate data with minimal loss of information. The information that is lost can be partly restored by generating a second principal component, and so on. In turn, the information from each individual principal component (PC) can be presented graphically in synthetic maps. These maps show peaks and troughs, which represent populations whose gene frequencies take extreme values compared to others in the studied area.Peaks and troughs usually, but not necessarily, connected by smooth gradients, called clines. Genetic clines can be generated in several ways: including adaptation to environment (natural selection), continuous gene flow between two initially different populations, or a demographic expansion into a scarcely populated environment with little initial admixture with pre-existing populations. Cavalli-Sforza connected these gradients with postulated pre-historic population movements based on known archaeological and linguistic theories. However, given that the time depths of such patterns are not known, “associating them with particular demographic events is usually speculative”.
Rewrite.
Luigi Luca Cavalli-Sforza performed studies on classical genetic markers to assess the geographic relationships between peoples in order to analyse DNA by proxy. Subsequently, the genetic distances between populations were calculated and phylogenetic trees were constructed that illustrated the genetic distances. These studies indicated relationships between geographically populations with modes and anti-modes connected generally by smooth gradients called clines.
This is all that is encyclopedic with regard to this page, the remainder of information belongs on a techniques page as it pertains to typing and phylogenetic techniques in general. For passages the need special emphasis please use footnotes.PB666 19:09, 21 July 2009 (UTC)
Studies using direct DNA analysis are now abundant use utilize mitochondrial DNA (mtDNA), the non-recombining portion of the Y chromosome (NRY) or autosomal DNA. MtDNA and NRY DNA share some similar features which have made them particularly useful in molecular anthropology. These properties include the direct, unaltered inheritance of mtDNA and NRY DNA from mother to offspring, and father to son, respectively, without the 'scrambling' effects of genetic recombination . We also presume that these genetic loci are not affected by natural selection , and that the major process responsible for changes in base pairs has been mutation (which can calculated). The smaller effective population size of the NRY and mtDNA enhances the consequences of drift and founder effect relative to the autosomes, making NRY and mtDNA variation a potentially sensitive index of population composition. However, these biologically plausible assumptions are nevertheless not concrete. For example, Rosser suggests that climactic conditions may affect the fertility of certain lineages. Even more problematic, however, is the underlying mutation rate used by the geneticists. They often use different mutation rates, and therefore studies are frequently arriving at vastly different conclusions. Moroever, NRY and mtDNA may be so susceptible to drift that some ancient patterns may have become obscured over time. Another implicit assumption is that population genealogies are approximated by allele genealogies. Barbujani points out that this only holds if population groups develop from a genetically monomorphic set of founders. However, Barbujani argues that there is no reason to believe that Europe was colonized by monomorphic populations. This would result in an overestimation of haplogroup age, thus falsely extending the demographic history of Europe into the Late Paleolithic rather than the Neolithic era. (See also Genetic drift, Founder effect, Population bottleneck.)
Here the question is do people who understand the complexity of human molecular anthropology produce alot of speculation or are they simply sitting back and waiting for better data to come that produces a non-speculative result. And if we are offering up speculation here what is its future value, here.PB666 19:46, 21 July 2009 (UTC)
Whereas Y-DNA and mtDNA haplogroups represent but a small component of a person’s DNA pool, autosomal DNA has the advantage of containing hundreds and thousands of examinable genetic loci, thus giving a more complete picture of genetic composition (eg see Seldin). However, descent relationships can only to be determined on a statistical basis because autosomal DNA undergoes recombination and is liable to the process of natural selection.
Genetic studies operate on numerous assumptions and suffer from usual methodological limitations such as selection bias and confounding. Furthermore, no matter how accurate the methodology, conclusions derived from such studies are ultimately compiled on the basis of how the author envisages their data fits with established archaeological or linguistic theories.
How does all of this pertain to the main? Is it a red flag on a bull's tail.PB666 19:42, 21 July 2009 (UTC)
Backward emphasis
Its interesting reading this article that the emphasis on genetics is almost the opposite of the genetic reliability of the markers used. S-Cs work is generally the least reliable, as it uses structure that are not discrete measures of molecular genetics, the Y chromosome which has all sorts of problem is treated as secondary importance, and the mtDNA which probably best represents the process gets a paragraph. Tishkoff's CD4 intron study is not mentioned at all, this was the first study showing some more recent african contribution at the molecular genetic level. HLA studies are not mentioned at all. There is way too much emphasis on old studies proxy methods that can have multiple interpretations, some of the conclusions fly against some of the more recent conclusions based on mtDNA and can be confirmed by HLA.PB666 19:55, 21 July 2009 (UTC)
References for Sub-Saharan Africa
- Genes peoples languages CS for genetic distances
- Cruciani; et al. (2004). "Phylogeographic Analysis of Haplogroup E3b (E-M215) Y Chromosomes Reveals Multiple Migratory Events Within and Out Of Africa".
{{cite journal}}: Cite journal requires|journal=(help); Explicit use of et al. in:|last=(help) - Bowcock; et al. (1991). "Drift, admixture, and selection in human evolution: A study with DNA polymorphisms".
{{cite journal}}: Cite journal requires|journal=(help); Explicit use of et al. in:|last=(help)
- King (2007). "Africans in Yorkshire? - the deepest-rooting clade of the Y phylogeny within an English genealogy".
{{cite journal}}: Cite journal requires|journal=(help) - Malyarchuk; et al. (2005). "African DNA Lineages in the Mitochondrial Gene Pool of Europeans".
{{cite journal}}: Cite journal requires|journal=(help); Explicit use of et al. in:|last=(help) - Malyarchuk; et al. (2008). "Reconstructing the phylogeny of African mitochondrial DNA lineages in Slavs".
{{cite journal}}: Cite journal requires|journal=(help); Explicit use of et al. in:|last=(help) - Halder; et al. (2008). "A panel of ancestry informative markers for estimating individual biogeographical ancestry and admixture from four continents: utility and applications".
{{cite journal}}: Cite journal requires|journal=(help); Explicit use of et al. in:|last=(help) - Auton; et al. (2009). "Global distribution of genomic diversity underscores rich complex history of continental human populations".
{{cite journal}}: Cite has empty unknown parameter:|1=(help); Cite journal requires|journal=(help); Explicit use of et al. in:|last=(help) - Reich (2008). "Principal component analysis of genetic data".
{{cite journal}}: Cite journal requires|journal=(help) - Gonzalez; et al. (2003). "Mitochondrial DNA Affinities at the Atlantic Fringe of Europe" (PDF).
{{cite journal}}: Cite journal requires|journal=(help); Explicit use of et al. in:|last=(help) - Gonzalez; et al. (2007). "Mitochondrial lineage M1 traces an early human backflow to Africa".
{{cite journal}}: Cite journal requires|journal=(help); Explicit use of et al. in:|last=(help) - Underhill; et al. (2001). "The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations" (PDF).
{{cite journal}}: Cite journal requires|journal=(help); Explicit use of et al. in:|last=(help)
Wapondaponda (talk) 00:29, 22 July 2009 (UTC)
- Martinez et al. (2007)
- Abu-Amero et al. (2007)
- Richards et al. (2003)
- Gonçalves et al. (2003)
- Pereira et al. (2000) Diversity of mtDNA lineages in Portugal: not a genetic edge of European variation. Ann Hum Genet; 64(Pt 6):491-506.
- Achilli et al. 2007, Malyarchuk et al. 2008, Gonzalez et al. (2003)
- Cruciani et al. 2004, Flores et al. 2004, Brion et al. 2005, Brion et al. 2004, Rosser et al. 2000, Semino et al. 2004, DiGiacomo et al. 2003
- Pritchard et al. (2000) Inference of Population Structure Using Multilocus Genotype Data. Genetics Society of America; 164(4):1567-87.
- Rosenberg et al. 2002, Wilson et al. 2001, Bauchet et al. 2007, Auton et al. 2009
Cite error: There are <ref group=note> tags on this page, but the references will not show without a {{reflist|group=note}} template (see the help page).